Abstract
Frequent outbreaks of novel coronaviruses (CoVs), highlighted by the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, necessitate the development of therapeutics that could be easily and effectively administered worldwide. The conserved mRNA-capping process enables CoVs to evade their host immune system and is a target for antiviral development. Nonstructural protein (nsp) 16 in complex with nsp10 catalyzes the final step of coronaviral mRNA capping through its 2′-O-methylation activity. Like other methyltransferases, the SARS-CoV-2 nsp10-nsp16 complex is druggable. However, the availability of an optimized assay for high-throughput screening (HTS) is an unmet need. Here, we report the development of a radioactivity-based assay for the methyltransferase activity of the nsp10-nsp16 complex in a 384-well format, kinetic characterization, and optimization of the assay for HTS (Z′ factor = 0.83). Considering the high conservation of nsp16 across known CoV species, the potential inhibitors targeting the SARS-CoV-2 nsp10-nsp16 complex may also be effective against other emerging pathogenic CoVs.
Keywords: COVID-19, nsp16, nsp10-nsp16 complex, SARS-CoV-2, methyltransferase
Introduction
The recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; also known as COVID-19) has posed a remarkable threat to global public health and the economy.1 Coronaviruses (CoVs) are major viral pathogens of humans and animals, including bats, livestock, and numerous wild animals.2 , 3 Among recognized CoV species,1 only seven are documented to infect humans. SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 cause severe symptoms in humans, while 229E, HKU1, OC43, and NL63 are associated with mild symptoms.2 , 4 CoVs belong to the family of Coronaviridae. Their genome is a single-stranded RNA, which encodes 16 functional nonstructural proteins (nsp1 to nsp16) and four main structural and accessory proteins.5 Most nonstructural proteins are involved in the synthesis and processing of CoV RNAs.6
Several endogenous 5′-mRNA cap structures are identified in mature eukaryotic cells that help recruit various cellular factors to support efficient translation and improve mRNA stability.7 , 8 Additionally, mRNA caps provide a molecular platform to differentiate between self and foreign mRNAs,9 which could result in the initiation of host immune responses when viral mRNAs are detected. In return, viruses have evolved an array of systems to combat these restrictions.10 , 11 Viral mRNAs that maintain both the 7-methylguanosine cap (N7-meGpppN; called Cap-0) and 2′-O-methylation of the first nucleotide (N7-meGpppN2′Ome; Cap-1) could stay viable like the host mRNAs.12 , 13 Particularly, CoVs generate mRNAs with a type 1 cap structure to avoid identification and activation of host defense mechanisms.13, 14, 15, 16 Coronaviral mRNA capping starts with the removal of 5′-γ-phosphate from nascent viral RNA by nsp13. A guanosine monophosphate is then attached to the 5′-diphosphate by an RNA guanylyltransferase to form GpppN-RNA. Subsequently, nsp14 methylates N7 of guanosine, giving rise to a Cap-0. Ultimately, Cap-0 is transformed into a doubly methylated (Cap-1) structure by nsp16.9 , 17
The importance of conserved nsp16 for the function and survival of CoVs has been documented in vivo and in vitro.13 , 16 , 18 , 19 Nsp16 is a member of the 2′-O-methyltransferase (MTase) family, catalyzing the transfer of a methyl group from S-adenosyl methionine (SAM) to RNA substrates.20 MTases are generally druggable with several highly selective and cell-active inhibitors of human MTases available.21 The in vitro 2′-O-MTase activity of nsp16 has been reported for feline-CoV, MERS-CoV, and SARS-CoV.22, 23, 24, 25 However, nsp16 was significantly active only when in complex with nsp10.23 Nsp16 alone is unstable, and nsp10-nsp16 complex formation is essential for its binding to SAM and RNA substrate.24 The crystal structure of nsp16 has only been determined in complex with nsp10.26 Several structures of nsp10-nsp16 from various CoV species are available as apo, and in complex with RNA substrate, SAM, or SAM analogs, which vastly enables the structure-based hit optimization.24 , 26, 27, 28, 29, 30 The Nsp10-nsp16 complex selectively binds and methylates longer CoV mRNAs and synthetic small RNAs with Cap-0.23 Moreover, SARS-CoV nsp10-nsp16 methylates N7-meGpppA-RNA but not N7-meGpppG-RNA, which provides some selectivity over the host mRNAs.24
These studies indicate that the conserved nsp10-nsp16 complex is essential for CoVs’ ability to mimic the host mRNAs needed for viral replication.12 , 13 , 23 Therefore, inhibition of nsp10-nsp16 complex activity could potentially hinder the pathogenesis of CoVs through eliciting a host immune response.13 , 15 , 16 However, the availability of an optimized assay suitable for high-throughput screening (HTS) is an unmet need. Here, we report the development and optimization of a scintillation proximity assay (SPA) for testing the RNA MTase activity of the nsp10-nsp16 complex, kinetic characterization, and HTS.
Materials and Methods
Reagents
Biotinylated RNA substrate (5′-N7-meGpppACCCCC-biotin) was synthesised by bioSYNTHESIS (Lewisville, Texas). Three hundred eighty-four- and 96-well Streptavidin PLUS High-Capacity FlashPlates, 3H-SAM, and 3H-biotin were from PerkinElmer (Waltham, MA). SAM, sinefungin, and S-adenosyl homocysteine (SAH) were from Sigma-Aldrich (St. Louis, MO). SAM2 Biotin-Capture Membrane was obtained from Promega (Madison, WI). All reaction buffers contained 0.4 U/μL RNaseOUT ribonuclease inhibitor (Invitrogen, Waltham, MA).
Protein Expression and Purification
Expression and purification of the SARS-CoV-2 nsp10-nsp16 complex was recently described.31 Briefly, nsp16 (S1-N298) and nsp10 (A1-Q139) were separately expressed in Escherichia coli BL21(DE3) RIL and purified to near homogeneity. The nsp10-nsp16 complex was prepared using the purified proteins in an 8 (nsp10) to 1 (nsp16) molar ratio; dialyzed in storage buffer containing 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.5 mM TCEP, and 5% glycerol; and flash frozen.
Optimization of nsp10-nsp16 Assay Conditions
The initial MTase reactions were performed in a buffer similar to the reported conditions for the SARS-CoV nsp10-nsp16 complex,23 with some modifications. Accordingly, 10 μL mixtures containing 50 mM Tris (pH 8.0), 1 mM MgCl2, 5 mM DTT, 2 µM RNA substrate, and 250 nM nsp10-nsp16 complex were prepared. The reactions were started by addition of 4 µM SAM (16% 3H-SAM). Reactions proceeded for 1 h and then were quenched by adding 10 μL of 7.5 M guanidine hydrochloride, followed by 60 μL of 20 mM Tris-HCl (pH 8.0). The reaction products were transferred into streptavidin-coated FlashPlates for scintillation counting using a TopCount instrument (PerkinElmer). Reaction mixtures were prepared in triplicate. For determining the optimum buffer pH, 50 mM Tris-HCl was used for generating the pH profile ranging from 6.5 to 9.0. The effects of various reagents such as salts, detergents, reducing agents, bovine serum albumin (BSA), EDTA, and DMSO were investigated through titration of each reagent in assay buffer at pH 7.5 and measuring their relative activity compared with the control (i.e., reactions without additive) using the SPA-based assay. The following buffer was chosen as the optimal reaction condition: 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 1.5 mM MgCl2, 0.01% Triton X-100, 0.01% BSA, and 5 mM DTT. All subsequent experiments were performed using this buffer condition. All reactions were performed at room temperature (23 °C).
Determination of Kinetic Parameters for nsp10-nsp16 Complex
For determining the kinetic parameters, reactions were carried out using the optimized buffer condition in triplicate in standard 96-well polypropylene plates. For each experiment, the concentration for one substrate (i.e., SAM or RNA) was varied, while the concentration of the second substrate was kept at near saturation (>3.5× K m). After starting the reaction by adding 3H-SAM, samples were taken at different time points, which were quenched by adding an equal volume of 7.5 M guanidine hydrochloride. The level of 3H-methylated RNA in each reaction was quantified by measuring the radioactivity level (counts per minute [CPM]) employing both a high-capacity biotin-capture membrane-based approach and an SPA-based method. For the membrane-based method, 12 μL of the quenched mixture was spotted on streptavidin-coated SAM2 biotin-capture membranes. The membranes were washed twice with 2M NaCl (2 min each), rinsed with water three times (2 min each), and allowed to fully air dry. Subsequently, the spotted squares were cut from the membranes and placed separately into scintillation vials containing scintillant solution, MicroScint-O (PerkinElmer). Finally, the 3H-methyl-RNA product was quantified using a Tri-Carb scintillation counter (PerkinElmer). In the SPA-based approach, the quenched mixtures were transferred into 96-well FlashPlates after adding 150 μL of 20 mM Tris-HCl (pH 8.0) to each well. To avoid saturation of beads in these plates with excess RNA substrate, only 6 μL of the quenched mixture was added to each well. 3H-Biotin at different concentrations was used as control. After overnight incubation, the level of 3H-methylated RNA was measured by scintillation counting. Afterward, the initial velocities were calculated from the linear portions of the reaction progression curves. The kinetic parameters were determined using Michaelis–Menten equation by GraphPad Software (La Jolla, CA). For clarity, when we report the activity of the protein complex as nmole/min/mg, “mg” refers to “mg of nsp16.” Since the complex is a 1:8 ratio of nsp16 to nsp10, the molarities of nsp16 and the nsp10-nsp16 complex are the same.
Z′-Factor Determination
The quality and robustness of the nsp10-nsp16 assay was verified by the standard Z′-factor determination.32 Optimized reaction mixtures containing 125 nM nsp10-nsp16 complex and 0.8 μM RNA were prepared in the presence or absence of 200 μM sinefungin in the 384-well format using an Agilent Bravo automated liquid handling robot. The final DMSO concentration was 1%. The reactions were started by addition of 1.7 μM SAM (30% 3H-SAM) and were incubated for 30 min at 23 °C. After measuring signal by the SPA-based method, the Z′-factor was calculated as previously described.32
Screening a Collection of Chemical Probes
The library of 76 epigenetic chemical probes was from the Structural Genomics Consortium (SGC; https://www.thesgc.org/chemical-probes/epigenetics). The compounds were screened at 50 μM with a final DMSO concentration of 1% in 125 nM nsp10-nsp16 complex, 0.8 μM RNA, and 1.7 μM SAM (30% 3H-SAM). Reactions containing 50 μM SAH and 1% DMSO were used as positive and negative controls, respectively. After 30 min of incubation, reactions were quenched and transferred into SPA plates, and the incorporated radioactivity was quantitated as described above. The final reaction mixture for screening the nsp10-nsp16 complex was 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 1.5 mM MgCl2, 0.01% Triton X-100, 0.01% BSA, 5 mM DTT, 125 nM nsp10-nsp16, 0.8 μM RNA, and 1.7 μM SAM (30% 3H-SAM). Supplemental Scheme 1 illustrates the screening workflow.
Sequence Analysis
Nsp16 protein sequences were taken from the CoV ORF1ab sequences accessible through the UniProt database. These sequences consisted of 229E (P0C6X1), HKU1 (P0C6X3), NL63 (P0C6X5), OC43 (P0C6X6), MERS-CoV (K9N7C7), SARS-CoV (P0C6X7), and SARS-CoV-2 (P0DTD1). The nsp16 sequences were aligned using Clustal Omega,33 and sequence similarities and secondary structure features were rendered by ESPript version 3.0.34 The conservation among these sequences was mapped onto the crystal structure of nsp10-nsp16 from SARS-CoV-2 (Protein Data Bank [PDB]: 6WKS) using Chimera version 1.14.35
Results
Assay Development and Optimization
The in vitro activity of the SARS-CoV-2 nsp10-nsp16 complex was tested by monitoring the transfer of 3H-SAM to the biotinylated N7-meGpppACCCCC RNA substrate. The methylated RNA product was captured using SPA plates followed by recording the changes in CPM. Initial tests at 250 nM nsp10-nsp16 complex, 2 μM RNA substrate, and 5 μM SAM indicated that the protein complex is active with a significant signal-to-noise ratio. The assay conditions were further optimized with respect to the pH of the buffer and the presence of several commonly used additives ( Fig. 1). The complex was most active at pH 7.5 and pH 8 (Fig. 1A). Using pH 7.5, the effects of other buffer components were investigated. Although NaCl over a wide range of concentrations (10–100 mM) reduced the enzyme activity by about 30%, KCl had little effect on nsp10-nsp16 complex activity up to 100 mM, and MgCl2 slightly increased the signal (Fig. 1B–D). However, a presence of Triton X-100 as low as 0.002% increased the signal by more than 20% (Fig. 1E). Tween-20 had a similar effect (Suppl. Fig. S1A). The reducing agents TCEP and DTT had no significant effect on enzyme activity (Fig. 1G,H). The presence of BSA at concentrations higher than 0.02% reduced the signal readout (Fig. 1F). EDTA at concentrations as low as 50 µM considerably reduced the activity (Suppl. Fig. S1B). Based on these observations, 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 1.5 mM MgCl2, 0.01% Triton X-100, 0.01% BSA, and 5 mM DTT were selected as the optimized buffer condition for SARS-CoV-2 nsp10-np16 complex MTase activity assays. Overall, the assay optimization led to a 70% increase in assay signal over the starting assay conditions (Suppl. Fig. S1C). The nsp10-nsp16 complex activity under the optimized conditions was not affected by DMSO up to 5% (Fig. 1I).
Figure 1.
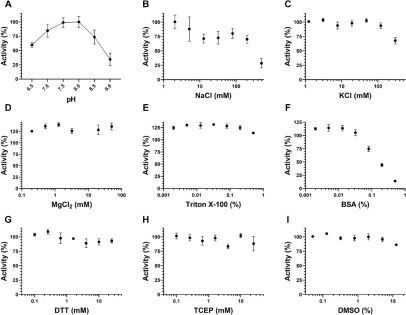
Assay optimization for SARS-CoV-2 nsp10-nsp16 complex activity. (A) The MTase activity of the nsp10-nsp16 complex was tested at various pH values. Tris-HCl at 50 mM was used to generate the pH gradient from 6.5 to 9.0. Using the optimal pH (7.5), the effects of (B) NaCl, (C) KCl, (D) MgCl2, (E) Triton X-100, (F) BSA, (G) DTT, (H) TCEP, and (I) DMSO were evaluated. Experiments were performed in triplicate (n = 3). Values are presented as mean ± standard deviation.
Kinetic Characterization
The kinetic parameters for the nsp10-nsp16 complex were determined using the optimized conditions. Initial assessment of the MTase activity at various concentrations of the nsp10-nsp16 complex indicated reaction linearity up to around 250 nM of the protein complex (Suppl. Fig. S1D). At 250 nM nsp10-nsp16 complex, using the membrane-based approach, apparent K m values of 1.7 ± 0.3 μM and 1.6 ± 0.4 μM were determined for SAM and RNA, respectively, with an apparent k cat of 15.9 ± 1.2 h–1 (Suppl. Fig. S2A–D). For determining the K m of SAM, the concentration of RNA was kept at 5.6 μM, whereas when assessing the K m of the RNA substrate, the SAM concentration was at 6.0 μM. To investigate if lowering the concentration of the nsp10-nsp16 complex is possible without nsp16 inactivation due to complex dissociation, the kinetic parameters for SAM and RNA were also determined at 125 nM nsp10-nsp16 complex. Using SPA, the linear initial velocities were used to calculate the kinetic parameters. For reactions at 125 nM enzyme, the linear first 5 min of the reactions ( Fig. 2A,B) was used to calculate the kinetic parameters (Fig. 2C,D). In these experiments, the concentration of the second substrates, SAM and RNA, were kept at 8.0 μM (Fig. 2C) and 5.0 μM (Fig. 2D), respectively. The apparent K m values of 1.0 ± 0.1 μM and 2.0 ± 0.2 μM for RNA substrate and SAM were determined, respectively (Fig. 2C,D). The apparent k cat value was 26.9 ± 0.3 h–1 (Fig. 2C–E). While only the first 20 min of the reactions is shown in Figure 2A,B, the extended range up to 50 min is presented in Supplemental Figure S2E,F. These data indicate that nsp10 and nsp16 stay in complex at a lower concentration and the integrity of the complex was not affected by further dilution of the protein complex. Therefore, all further assays were performed at 125 nM nsp10-nsp16 complex. The N7-unmethylated biotinylated RNA substrate, which was used as a control, showed almost no activity under similar assay conditions (Suppl. Fig. S1E).
Figure 2.
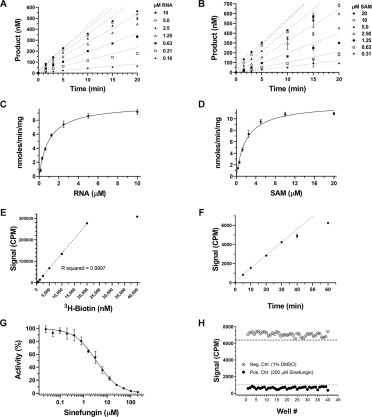
Kinetic parameter determination for the nsp10-nsp16 complex. The initial velocities were determined at 125 nM nsp10-nsp16 complex using (A) various concentrations of RNA (as indicated on the plot) and a fixed SAM concentration (8 µM) and (B) varying concentrations of SAM and a fixed RNA concentration of 5 µM under the optimized condition. For the first 5 min, linear initial velocities from A and B were used to calculate the Km values for (C) RNA substrate and (D) SAM. The data on running the reactions up to 50 min are presented in Supplemental Figure S2E,F. (E) A standard 3H-biotin titration curve was generated and used for calculating the kcat values. (F) The linearity of nsp10-nsp16 activity over time using the optimized buffer and final screening conditions (0.8 μM RNA, 1.7 μM SAM, and 125 nM nsp10-nsp16 complex) was confirmed for 30 min. (G) Sinefungin inhibited nsp10-nsp16 activity with an IC50 of 3.4 ± 0.4 μM (Hill Slope, –0.9). All experiments (A–G) were performed in triplicate (n = 3). (H) The Z′-factor was determined in the presence (filled black circles) and absence (empty circles) of 200 μM sinefungin using screening conditions. The dashed and dotted lines represent 3 standard deviations from the mean for each control group.
Assessment of HTS Amenability
To assess the quality of the assay for HTS, first the linearity of the reaction over time at slightly below the K m of each substrate (0.8 μM RNA and 1.7 μM SAM) was assessed ( Table 1, Fig. 2F). The data revealed that the reaction was linear for at least 30 min. Using this assay condition, it was shown that sinefungin inhibited nsp10-nsp16 activity with an IC50 of 3.4 ± 0.4 μM (Hill Slope, –0.9) (Fig. 2G). Subsequently, the quality and robustness of the developed assay for HTS was analyzed. For screening in a 384-well format, a Z′-factor of 0.83 was attained (Fig. 2H). The optimized assay was then employed to screen a panel of 76 epigenetic chemical probes (Suppl. Fig. S3), which included more than 20 potent MTase inhibitors (Suppl. Table S1). At a final compound concentration of 50 μM, none of these highly selective compounds significantly inhibited (>26%) the activity of the nsp10-nsp16 complex, while SAH (IC50 of 5.9 ± 0.6 μM; Suppl. Fig. S1F) reduced the activity of nsp10-nsp16 by >90% at 50 μM.
Table 1.
Kinetic Characterization of the nsp10-nsp16 Complex.
| Experiment | nsp16 (nM) | Substrate | Kmapp (µM) | kcat (h–1) |
|---|---|---|---|---|
| 1 | 250 | SAM RNA |
1.7 ± 0.3 1.6 ± 0.4 |
15.9 ± 1.2 |
| 2 | 125 | SAM RNA |
2.0 ± 0.2 1.0 ± 0.1 |
26.9 ± 0.3 |
Values are presented as mean ± SD (n = 3).
Discussion
As the fight against COVID-19 continues, several vaccines against SARS-CoV-2 have been made available to the public. However, administering these vaccines requires very specific handling protocols, such as extremely low storage temperature for some, which may not be easily achievable in many countries. Even if all conditions are met, it will take many months to complete the vaccination. In addition, these vaccines may not be effective for fast mutating CoVs. This necessitates antiviral development.36 The 2′-O-MTase nsp16 has been proposed as an appealing target for the development of anticoronaviral therapeutics.8 , 11 , 28 , 37 Deletion of the SARS-CoV nsp16 coding region resulted in a blockade of viral RNA synthesis,18 and nsp16 mutants have shown a strong attenuation in infected mice.19 It has been suggested that the nsp10-nsp16 complex, through its mRNA-capping activity, helps the CoVs to evade the host immune system.15 Therefore, any interruption in the activity of nsp10-nsp16 could hinder the pathogenesis of CoVs by eliciting an immune response.13 , 15 , 16 Inhibition of nsp10-nsp16 complex MTase activity by SAH (the product of the reaction), sinefungin (a SAM analog), and aurintricarboxylic acid has been reported.22 , 23 , 38 However, potent and cell-permeable nsp10-nsp16 inhibitors are yet to be developed. The availability of activity-based HTS assays would greatly enable drug discovery. The activity of the SARS-CoV nsp10-nsp16 complex has previously been assessed using a filter binding-based assay.22 Most recently, an HTS RNA displacement assay has been reported for the SARS-CoV-2 nsp10-nsp16 complex that will detect RNA-competitive inhibitors.31 Overall, the nsp10-nsp16 complex activity assays reported to-date are low throughput.22, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 , 38
Here we reported development of a radioactivity-based assay for screening the SARS-CoV-2 nsp10-nsp16 complex in a 384-well format. Since around 10-fold molar excess of nsp10 is required for the maximum in vitro MTase activity of nsp16,23 a 1:8 ratio of nsp16 to nsp10 was chosen to ensure a near-maximum activity of the complex. The kinetic parameters for the MTase activity of the nsp10-nsp16 complex are presented here for the first time. The K m values of SAM and RNA were determined to be 2.0 ± 0.2 μM and 1.0 ± 0.1 μM, respectively. The ITC K d values of 5.59 ± 1.15 μM and 1.21 ± 0.41 μM for SAM and RNA, respectively, were previously reported for the nsp10-nsp16 complex from SARS-CoV.24 The apparent k cat value for the SARS-CoV-2 nsp10-nsp16 complex is 26.9 ± 0.3 h–1. Such low k cat values are not uncommon among RNA39 , 40 or protein MTases.41 The IC50 values for SAH and sinefungin determined in this study were in the same range as values previously reported for the SARS-CoV and MERS-CoV nsp10-nsp16 complexes.22 , 23 Testing a subset of potent and selective chemical probes for human MTases did not significantly inhibit the activity of the nsp10-nsp16 complex, indicating that the assay has a very low rate of false positives and is well suited for HTS. Unlike RNA displacement assays, this MTase activity assay is suitable for detecting both SAM- and RNA-competitive inhibitors.
The available evidence indicates that many other CoVs currently in various animals are preadapted to likely infect humans at some point in time in the future and cause new pandemics.42, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44 Considering the natural diversity of CoVs across the globe1 and the close interactions of humans with wild and domesticated animals, these future pandemics may not be prevented by the current vaccines.36 This further highlights the importance of developing potent inhibitors against coronaviral proteins that are conserved across this family of viruses to develop pan-CoV therapeutics. Nsp16 is highly conserved across the CoV family,12 and the available structures from several coronaviral species also reveal a high degree of structural conservation.24 , 26, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 , 30 For example, SARS-CoV-2 nsp16 shows a minimum sequence identity of 57.05% with the other pathogenic CoVs ( Fig. 3). Mapping this sequence alignment on the nsp10-nsp16 structure ( Fig. 4) demonstrates the conservation of SAM- and RNA-binding pockets across CoV species. Therefore, inhibitors targeting the active site of nsp10-nsp16 may prove to be effective against other emerging and reemerging CoV strains. The radioactivity-based assay reported here will be an enabling tool toward developing such pan-inhibitors of nsp10-nsp16 MTase activities and possibly future pan-CoV therapeutics.
Figure 3.
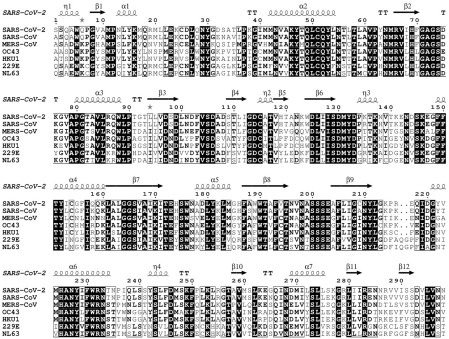
Sequence alignment of nsp16 from pathogenic CoVs. Amino acid sequences of nsp16 from seven pathogenic CoVs (HKU1, NL63, OC43, 299E, MERS-CoV, SARS-CoV, and SAR-CoV-2) were aligned using Clustal Omega, and sequence similarities and secondary structure features were rendered by ESPript 3.0. The crystal structure of SAR-CoV-2 nsp10-nsp16 (PDB: 7JHE) was employed for extracting the secondary structure information. SARS-CoV-2 nsp16 shows 57.05%, 58.39%, 66.11%, 63.76%, 66.11%, and 93.29% sequence identity to 299E, NL63, OC43, HKU1, MERS-CoV, SARS-CoV, respectively.
Figure 4.
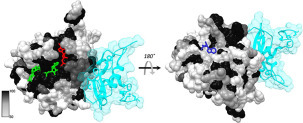
Sequence conservation of nsp16 across CoV species. The sequence conservation shown in Figure 3 is mapped onto the crystal structure of nsp10-nsp16 from SARS-CoV-2 (PDB: 6WKS). The structure was rendered by the percentage conservation of amino acid residues across the nsp16 from seven CoVs currently known to infect humans (darker colors represents a higher degree of conservation, with black being the highest). The nsp10 subunit is shown in transparent cyan. SAM is shown with a red stick, RNA is in green, and adenosine is represented by a blue stick.
In conclusion, the HTS assay we developed for assessing the activity of the SARS-CoV-2 nsp10-nsp16 complex using an SPA-based method provides a robust and sensitive tool for screening large libraries of compounds and is suitable for identifying inhibitors with different mechanisms of inhibition. It can also be employed as an orthogonal method for reevaluating potential inhibitors identified through other biochemical, biophysical, or cellular screening methods. Considering the critical role of the nsp10-nsp16 complex in coronaviral pathogenesis and the highly conserved nature of the nsp10-nsp16 complex across CoV species, the identified inhibitors may prove effective against other pathogenic CoVs, preventing future pandemics.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
We thank Dr. Aled Edwards and Dr. Cheryl Arrowsmith for continued support, and Dr. Peter Brown for critical review of the manuscript.
Supplemental material is available online with this article.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the University of Toronto COVID-19 Action Initiative-2020, Takeda California Inc., and COVID-19 Mitacs Accelerate postdoctoral awards to A.K.Y and S.P. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie, Bayer AG, Boehringer Ingelheim, Genentech, Genome Canada through Ontario Genomics Institute (OGI-196), the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510), Janssen, Merck KGaA (aka EMD in Canada and the United States), Pfizer, Takeda and the Wellcome Trust (106169/ZZ14/Z).
Supplemental Material
Supplemental Material
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K.G., Rambaut A., Lipkin W.I., et al. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y., Liu Q., Guo D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., Muth D., Niemeyer D., et al. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong S., Sun J., Mao Z., et al. A Guideline for Homology Modeling of the Proteins from Newly Discovered Betacoronavirus, 2019 Novel Coronavirus (2019-nCoV) J. Med. Virol. 2020;92:1542–1548. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuichi Y., Shatkin A.J. Viral and Cellular mRNA Capping: Past and Prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong H., Fink K., Zust R., et al. Flavivirus RNA Methylation. J. Gen. Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 9.Decroly E., Ferron F., Lescar J., et al. Conventional and Unconventional Mechanisms for Capping Viral mRNA. Nat. Rev. Microbiol. 2011;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramanathan A., Robb G.B., Chan S.H. mRNA Capping: Biological Functions and Applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decroly E., Canard B. Biochemical Principles and Inhibitors to Interfere with Viral Capping Pathways. Curr. Opin. Virol. 2017;24:87–96. doi: 10.1016/j.coviro.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menachery V.D., Debbink K., Baric R.S. Coronavirus Non-Structural Protein 16: Evasion, Attenuation, and Possible Treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devarkar S.C., Wang C., Miller M.T., et al. Structural Basis for m7G Recognition and 2′-O-Methyl Discrimination in Capped RNAs by the Innate Immune Receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai M.M., Patton C.D., Stohlman S.A. Further Characterization of mRNA’s of Mouse Hepatitis Virus: Presence of Common 5′-End Nucleotides. J. Virol. 1982;41:557–565. doi: 10.1128/jvi.41.2.557-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daffis S., Szretter K.J., Schriewer J., et al. 2′-O Methylation of the Viral mRNA Cap Evades Host Restriction by IFIT Family Members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zust R., Cervantes-Barragan L., Habjan M., et al. Ribose 2′-O-Methylation Provides a Molecular Signature for the Distinction of Self and Non-Self mRNA Dependent on the RNA Sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Guo D. Molecular Mechanisms of Coronavirus RNA Capping and Methylation. Virol. Sin. 2016;31:3–11. doi: 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almazan F., Dediego M.L., Galan C., et al. Construction of a Severe Acute Respiratory Syndrome Coronavirus Infectious cDNA Clone and a Replicon to Study Coronavirus RNA Synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menachery V.D., Yount B.L., Jr., Josset L., et al. Attenuation and Restoration of Severe Acute Respiratory Syndrome Coronavirus Mutant Lacking 2′-O-Methyltransferase Activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snijder E.J., Bredenbeek P.J., Dobbe J.C., et al. Unique and Conserved Features of Genome and Proteome of SARS-Coronavirus, an Early Split-Off from the Coronavirus Group 2 Lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheer S., Ackloo S., Medina T.S., et al. A Chemical Biology Toolbox to Study Protein Methyltransferases and Epigenetic Signaling. Nat. Commun. 2019;10:19. doi: 10.1038/s41467-018-07905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aouadi W., Blanjoie A., Vasseur J.J., et al. Binding of the Methyl Donor S-Adenosyl-l-Methionine to Middle East Respiratory Syndrome Coronavirus 2′-O-Methyltransferase nsp16 Promotes Recruitment of the Allosteric Activator nsp10. J. Virol. 2017;91 doi: 10.1128/JVI.02217-16. e02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvet M., Debarnot C., Imbert I., et al. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Su C., Ke M., et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decroly E., Imbert I., Coutard B., et al. Coronavirus Nonstructural Protein 16 Is a cap-0 Binding Enzyme Possessing (Nucleoside-2′O)-Methyltransferase Activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decroly E., Debarnot C., Ferron F., et al. Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan T., Arya S., Chan S.H., et al. Structural Basis of RNA Cap Modification by SARS-CoV-2. Nat. Commun. 2020;11:3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosas-Lemus M., Minasov G., Shuvalova L., et al. High-Resolution Structures of the SARS-CoV-2 2′-O-Methyltransferase Reveal Strategies for Structure-Based Inhibitor Design. Sci. Signal. 2020;13:eabe1202. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S., Chen H., Ye F., et al. Crystal Structure of SARS-CoV-2 nsp10/nsp16 2′-O-Methylase and Its Implication on Antiviral Drug Design. Signal Transduct. Target Ther. 2020;5:131. doi: 10.1038/s41392-020-00241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krafcikova P., Silhan J., Nencka R., et al. Structural Analysis of the SARS-CoV-2 Methyltransferase Complex Involved in RNA Cap Creation Bound to Sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perveen S., Khalili Yazdi A., Devkota K.; et al. A High-Throughput RNA Displacement Assay for Screening SARS-CoV-2 nsp10-nsp16 Complex toward Developing Therapeutics for COVID-19. SLAS Discov.2021. DOI: 10.1177/2472555220985040. [DOI] [PMC free article] [PubMed]
- 32.Zhang J.H., Chung T.D., Oldenburg K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 33.Sievers F., Wilm A., Dineen D., et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert X., Gouet P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Tse L.V., Meganck R.M., Graham R.L., et al. The Current and Future State of Vaccines, Antivirals and Gene Therapies against Emerging Coronaviruses. Front. Microbiol. 2020;11:658. doi: 10.3389/fmicb.2020.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferron F., Decroly E., Selisko B., et al. The Viral RNA Capping Machinery as a Target for Antiviral Drugs. Antiviral Res. 2012;96:21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barral K., Sallamand C., Petzold C., et al. Development of Specific Dengue Virus 2′-O- and N7-Methyltransferase Assays for Antiviral Drug Screening. Antiviral Res. 2013;99:292–300. doi: 10.1016/j.antiviral.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Blazer L.L., Li F., Kennedy S., et al. A Suite of Biochemical Assays for Screening RNA Methyltransferase BCDIN3D. SLAS Discov. 2017;22:32–39. doi: 10.1177/1087057116666276. [DOI] [PubMed] [Google Scholar]
- 40.Li F., Kennedy S., Hajian T., et al. A Radioactivity-Based Assay for Screening Human m6A-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. J. Biomol. Screen. 2016;21:290–297. doi: 10.1177/1087057115623264. [DOI] [PubMed] [Google Scholar]
- 41.Eram M.S., Kuznetsova E., Li F., et al. Kinetic Characterization of Human Histone H3 Lysine 36 Methyltransferases, ASH1L and SETD2. Biochim. Biophys. Acta. 2015;1850:1842–1848. doi: 10.1016/j.bbagen.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Hu B., Zeng L.P., Yang X.L., et al. Discovery of a Rich Gene Pool of Bat SARS-Related Coronaviruses Provides New Insights into the Origin of SARS Coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menachery V.D., Yount B.L., Jr., Sims A.C., et al. SARS-Like WIV1-CoV Poised for Human Emergence. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morens D.M., Fauci A.S. Emerging Pandemic Diseases: How We Got to COVID-19. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material


