Figure 1.
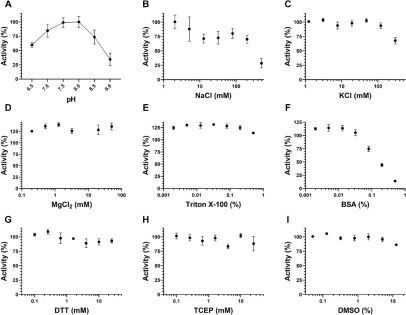
Assay optimization for SARS-CoV-2 nsp10-nsp16 complex activity. (A) The MTase activity of the nsp10-nsp16 complex was tested at various pH values. Tris-HCl at 50 mM was used to generate the pH gradient from 6.5 to 9.0. Using the optimal pH (7.5), the effects of (B) NaCl, (C) KCl, (D) MgCl2, (E) Triton X-100, (F) BSA, (G) DTT, (H) TCEP, and (I) DMSO were evaluated. Experiments were performed in triplicate (n = 3). Values are presented as mean ± standard deviation.
