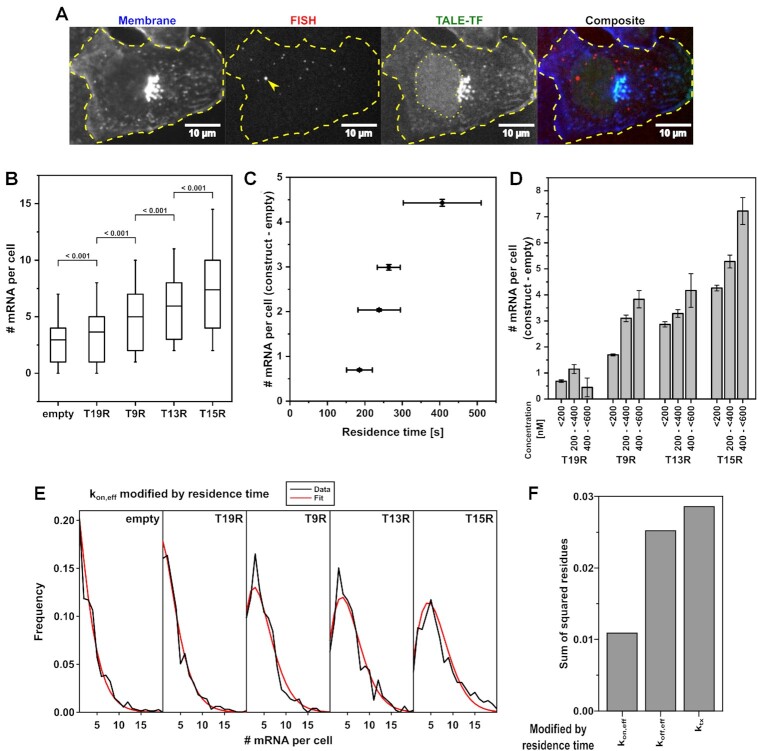
Figure 3.
TF residence time and concentration stimulate transcription activation. (A) Example images of the smFISH methodology: Lectin-FITC-stained cell membrane (blue; contrast 0 3000), smFISH of specific mRNA transcripts and nascent sites of transcription (arrow) (red; z-projection; contrast 2 40), TMR-stained HaloTag-TALE-TF (green; contrast 0 500) and composite image. Dashed line indicates the cell outline. Dotted line indicates the nucleus. The bright aggregate visualized by membrane stain and seen in the TALE-TF channel due to bleed-through probably corresponds to endoplasmic reticulum or Golgi apparatus. (B) mRNA distribution in cells expressing no TALE-TF (empty) and in cells expressing the different TALE-TF with < 300 nM. Cell numbers: N = 752 (empty); N = 888 (T19R); N = 690 (T9R); N = 664 (T13R); N = 790 (T15R). Mean (line), 25th/75th percentile (box) and 10th/90th percentile (whiskers) define features of box plot. Significance was determined with Wilcoxon–Mann–Whitney two-sample rank test. (C) Mean mRNA numbers from B) of cells expressing TALE-TF subtracted with the mean mRNA number of cells expressing no TALE-TF plotted versus TALE-TF residence time. Error bars denote s.e.m. for mean mRNA numbers and resampling error estimation for residence times. (D) Mean mRNA numbers of cells expressing TALE-TF at the indicated concentration subtracted with the mean mRNA number of cells expressing no TALE-TF. Cell numbers (<200 nM, 200 to <400 nM, 400 to <600 nM): T19R N = 655, 375, 113; T9R N = 509, 282, 102; T13R N = 478, 256, 33; T15R N = 580, 365, 155). Error bars denote s.e.m. (E) mRNA distributions of TALE-TFs together with the distribution inferred by BIRD for the case where TF residence time modifies kon,eff of the gene. (F) Comparison of mRNA distributions inferred by BIRD with the measured mRNA distributions of TALE-TFs by means of the lowest sum of squared residues in cases were TF residence time modifies kon,eff, koff,eff or ktx.