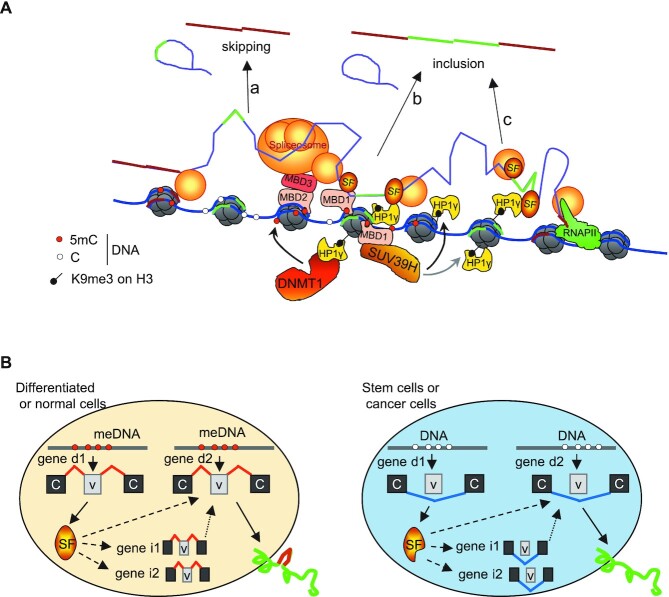
Figure 10.
Models for the roles of DNA methylation in the modulation of alternative splicing. (A) Absence of DNMT1/3b decreases CpG methylation (white circle), inhibits MBD recruitment, prevents the HP1γ spreading on the gene body, and favors the skipping of variant exons (A). DNA methylation (5mC) can be recognized by MBD proteins that may help regulate RNA alternative splicing by promoting S5p-mediated RNAPII pausing, by interacting with the spliceosome and/or splicing factors (SF), or by promoting HP1γ recruitment on chromatin via H3K9me3 provided by SUV39H1 (B). In the absence of CpG, high levels of H3K9me3 still favor the local recruitment of HP1γ (C) without counteracting the impact of the 5mC defect on splicing. (B) At directly impacted genes (gene d), some splicing factors, such as TRA2Aa/b, might act as a DNA methylation sensor by expressing specific isoforms which promote alternative splicing decision on genes indirectly regulated (genes i) and also on genes directly regulated (genes d) by DNA methylation. This last situation creates a positive loop reinforcing the modulation of alternative splicing by local 5mC levels.