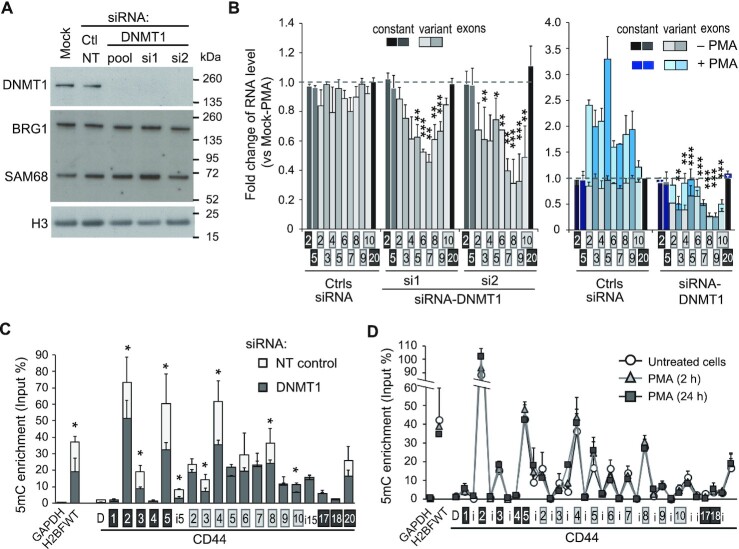
Figure 4.
CD44 variant exon levels depend on DNA methylation in DNMT1 depleted HeLa cells. (A, B) HeLa cells were transfected with two individual siRNAs or a commercially available pool of 3 siRNAs targeting DNMT1,or control siRNAs, either targeting GAPDH or non-targeting (NT). The cells were treated with or without PMA for 2 h before harvesting. (A) Western blot analysis of cell extracts using the indicated antibodies (see Supplementary Figure S4B for PMA-treated cell extract). (B) RT-qPCR analysis with primers aligning on the indicated exons. Relative RNA levels normalized to the RPLP0 gene as a reference, were used to calculate the fold change relative to the mock untreated cells (set to 1, dashed line). Statistical significance of the exonic differential levels upon DNMT1 depletion (without PMA; left panel) was calculated based on 4 independent experiments using Student's test (two-tailed), with P < 0.05 (*), P < 0.01 (**), P < 0.001 (***). The right panel shows the effect of PMA (blue bars) in these conditions of transfection with the DNMT1 siRNAs pool. Constitutive exons showed no significant difference between NT siRNA transfected cells (Ctrls) and mock transfected cells. The variant exons were increased by PMA in the presence of DNMT1. (C, D) CD44 intragenic DNA methylation is dependent on DNMT1 expression and independent of PKC activation. (C) DNA from HeLa cells transfected with the pool of DNMT1 siRNAs were analyzed by methylated DNA immunoprecipitation (MeDIP) using the 3D33 antibodies directed against methylated DNA (5mC) or non-immune IgG as negative control. The levels of 5mC were expressed as a percentage of the input DNA quantities for each of the qPCR primers covering the CD44 gene : intronic sequences (i) are indicated by white boxes, constant exons are indicated by black boxes and variant exons by light grey boxes, D is a distal sequence upstream to the TSS. The germline-specific H2BWT gene was used as a positive control for DNA methylation in the somatic HeLa cells, while the GAPDH promoter, a non-methylated CpG-rich region, was used to evaluate the background signal. The control IgG was at least 100 times less enriched and is not represented. Data are the average (±dev.) of 3 independent experiments. (D) HeLa cells were treated with or without PMA for the indicated time and the relative enrichment in 5mC was evaluated by MeDIP as in panel C.