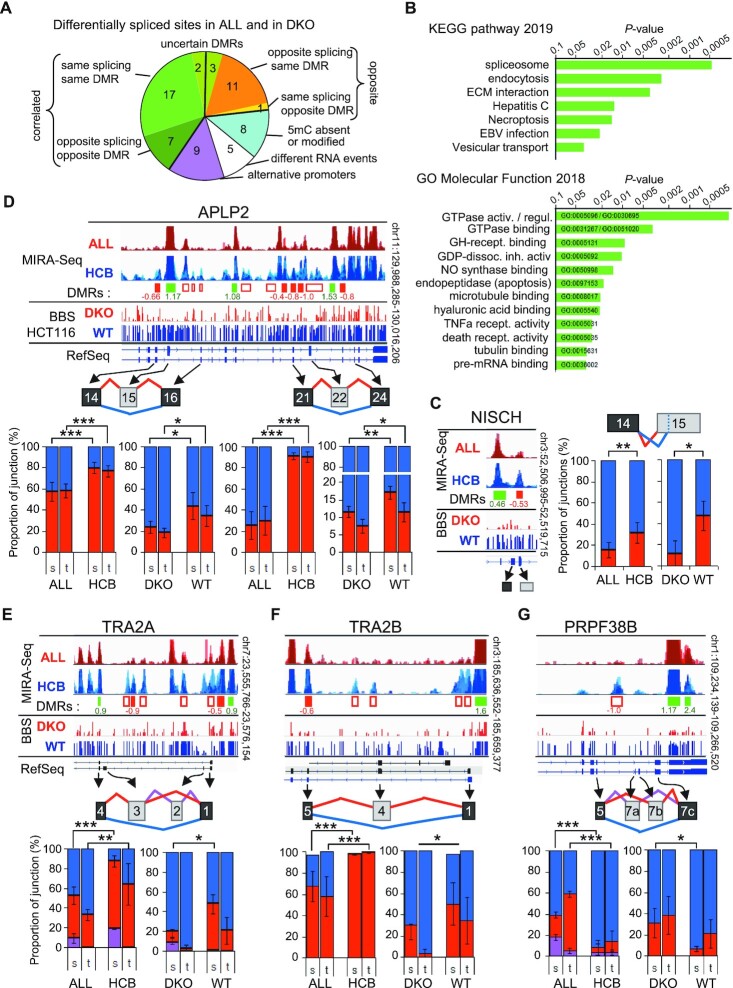
Figure 8.
A few differentially spliced genes may be an indication of DNA methylation and its biological relevance. (A) Differentially spliced genes from DKO (Supplementary Table S2) and from ALL (Supplementary Table S4) were compared to identify common splicing sites. RNA events were categorized as indicated. (B) Pathway analysis performed using Enrichr (127) on the 23 differentially spliced genes correlated with changes in DNA methylation from panel A (green quarters). (C–G). Examples of genes with common splicing events in DKO versus WT HCT116 cells and in ALL versus HCB pre-B cells. The overlaid MIRA-seq tracks set at the same scale, for ALL and HCB samples spanning the region containing the regulated cassette exon (in grey boxes). Changes in the DNA methylation levels were evaluated, and displayed as decreased (red boxes) or increased (green boxes) DMRs below. When found to be significant (P < 0.05) after normalization and Bonferroni's correction for multitesting, log2 fold change is indicated and boxes are colored. In addition, empty boxes indicates obvious DMRs. Below, the Methyl-seq (BBS) from DKO and WT HCT116 cells is displayed above genome view of the RefSeq genes. The graphs are the quantification of the RNA alternative splicing of each indicated variant exons detected with high confidence P(|dPSI| > 0.2) > 0.95 by MAJIQ. For each sample, counts of reads covering the indicated junctions were used to calculate the proportion of each junction involving the source exons (s) or the target exons (t), and represented as histogrammes of percentage of inclusion (red and pink) and skipping (blue). The six RNA-seq samples (APLP2), or the polyA+ RNA samples (NISCH, TRA2A, TRA2B, PRPF38B) of DKO and HCT116 cells were used to calculate the averages (± dev.) and the Student's t test (one-tailed): P < 0.05 (*), P < 0.01(**), P < 0.001 (***). The eight HCB samples and the 12 ALL samples selected in Supplementary Figure S6A and S6B, were used for the same calculation.