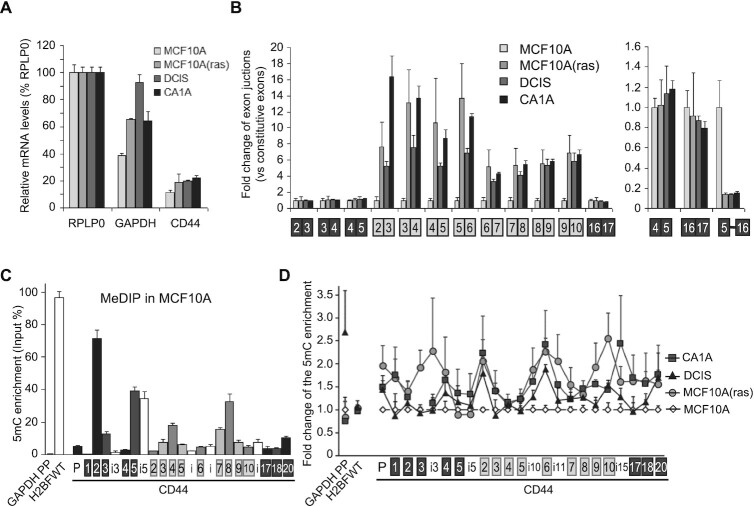
Figure 9.
DNA methylation increases with the inclusion of CD44 variant exons in tumorigenic-derived MCF10A-cell lines. (A, B) RNAs extracted from the cell lines were subjected to RT-qPCR using primers specific to the indicated regions. (A) The CD44 mRNA level were the average of constitutive exons relative to RPLP0 used as an unmodified reference gene. (B) Left, levels of the variant exons (grey boxes) were normalized to the average of constitutive exons (black) and expressed as a fold change relative to the levels in MCF10A (set to one for each primer pair). Right; levels of the CD44 isoform skipping the variant exons (meaning the C5 to C16C17 exon junction). (C, D) MeDIP assays performed on purified DNA from MCF10A using the 3D33 antibody directed against methylated DNA or non-immune IgG as negative control. Enrichment in 5mC is shown relative to the DNA amount in each input sample. The levels of DNA methylation in MCF10A parental cells is shown in C. The control IgG was at least 100 times less enriched and is not represented. (D) Enrichment in meDNA in other MCF10A-transformed cell lines relative to the levels in MCF10A at indicated loci. Data are the average (±dev.) of two independent experiments with triplicates used for qPCR. The statistical analysis is in Table 1.