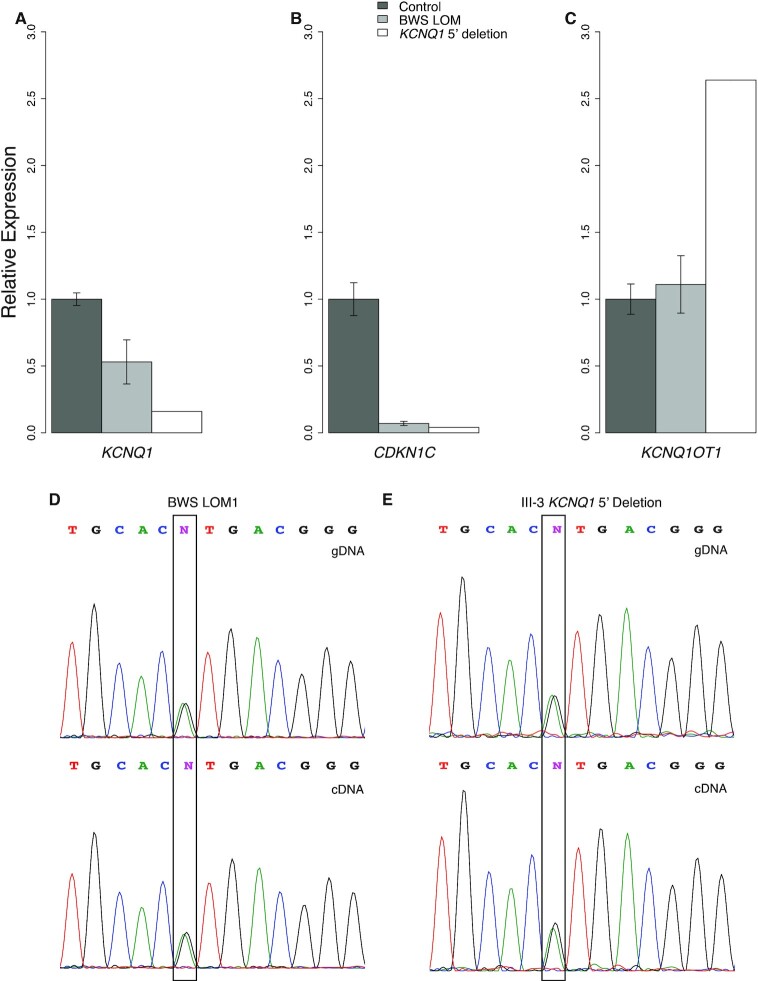
Figure 3.
Expression of KCNQ1, CDKN1C, and KCNQ1OT1 imprinted genes within IC2. Relative expression of IC2 imprinted genes was assessed by qRT-PCR and normalized to ACTB and RPLP0 levels. For LOM, N = 3; for control, N = 3. Error bars represent CI. (A) Expression of maternally-expressed KCNQ1 in placenta is reduced in BWS LOM cells and is further disrupted in III-3 cells carrying the maternal allele deletion relative to control samples from BWS LOM siblings. (B) Expression of maternally-expressed CDKN1C in fibroblasts is similarly aberrantly repressed in both BWS LOM and III-3 cells relative to unrelated non-BWS controls. (C) Expression of paternally-expressed KCNQ1OT1 in fibroblasts is increased in III-3 cells carrying the KCNQ1 5′ deletion relative to unrelated non-BWS controls. (D) Sanger sequencing trace of a KCNQ1OT1 SNP, rs231362 designated by the black box, in genomic (top) and complementary (bottom) DNA isolated from BWS LOM1 fibroblasts demonstrates biallelic expression. (E) Sanger sequencing trace of the boxed rs231362 SNP in genomic (top) and complementary (bottom) DNA isolated from III-3 KCNQ1 5′ deletion fibroblasts demonstrates biallelic expression.