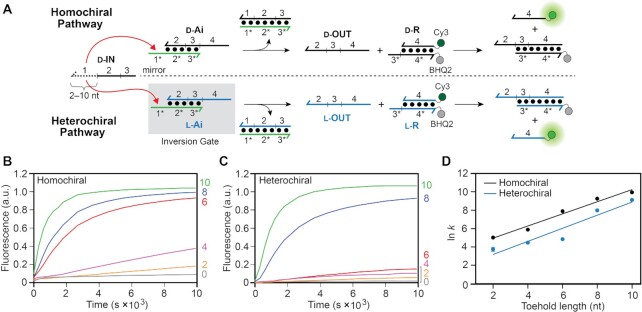
Figure 2.
Strand displacement from a PNA–DNA heteroduplex is dependent on toehold length and stereochemistry. (A) Schematic illustration of the strand displacement reaction system depicting both the homo- and heterochiral pathways. d-DNA is shown in black, l-DNA is shown in blue, and PNA is shown in green throughout the text. The half-arrow denotes the C-terminus of the PNA strand. The sequences of all strands are depicted in Supplementary Figure S1 and Supplementary Table S1. (B, C) Fluorescence monitoring (Cy3) of the homochiral (B) and heterochiral (C) reaction pathways initiated with inputs (d-IN) having toehold domains varying in length from 0–10 nucleotides (nt). The length of the toehold is indicated on the right y-axis. The reactions depicted contained 150 nM d-IN, 100 nM d/l-Ai, 300 nM d/l-R, 300 mM NaCl, 1 mM EDTA, and 10 mM Tris (pH 7.6) and were carried out at 37°C. Fluorescence in all figures is reported in units such that 1.0 is the fluorescence of the maximally activated reporter control and 0.0 is the background of the quenched reporter complex (d/l-R). (D) Semilogarithmic plot showing the exponential dependence of calculated rate constants on toehold length. All calculated rate constants are listed in Supplementary Table S2.