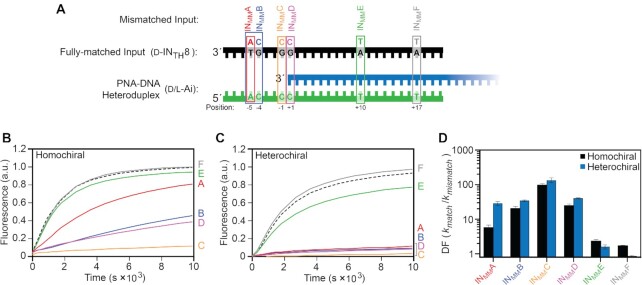
Figure 3.
The position of a mismatch affects the rate of strand displacement from PNA–DNA heteroduplexes. (A) Schematic of the mismatched inputs used in this study. The identity of the mismatched nucleotide relative to the fully matched input (d-INTH8) is shown above the strand. (B, C) Fluorescence monitoring (Cy3) of the homochiral (B) and heterochiral (C) reaction pathways initiated with different mismatched inputs in (A). The identity of the mismatched input (INMMA–F) is indicated on the right y-axis. The reaction initiated with the fully-matched input (d-INTH8) is shown as a black dotted line. Reactions depicted here were carried out as described in Figure 2. (D) Kinetic discrimination factors (DF = kmatch/kmismatch) of hetero- and homo-chiral reactions towards inputs having different mismatches, where kmatch and kmismatch are the calculated rate constants for a fully-matched and mismatched input, respectively. Error bars represent standard deviation from three independent experiments.