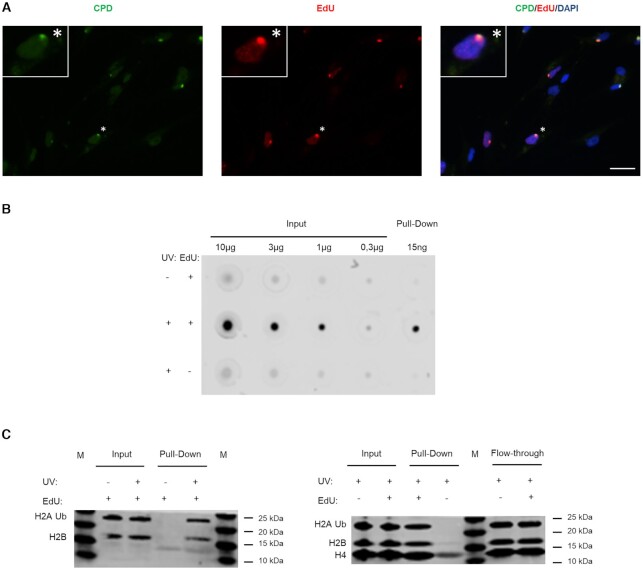
Figure 2.
Specific labelling and isolation of the UDS-associated chromatin. (A) 1BR.3 fibroblasts were locally irradiated with 100 J/m2 UVC using 8 μm pore filters and incubated for 4 h in no serum medium containing EdU and HU. The coverslips were incubated with Alexa-azide for labelling the incorporated EdU, and with anti-CPD. Scale bar: 25 μm. (B) VH10 fibroblasts were synchronized for aniFOUND and were UVC- or mock-irradiated with 30 J/m2. They were kept in the presence of HU and in the presence/absence of EdU for 4 h. Next, cells were subjected to aniFOUND and the DNA from the isolated material (as well as from input) was extracted, quantified and immobilized on a nitrocellulose membrane. The membrane was incubated with Streptavidin-Alexa for detecting the incorporated EdU. C1 Dynabeads were used for this experiment. The blot was cropped to advance clarity. (C) Fibroblasts were synchronized for aniFOUND and then were either UVC- or mock-irradiated with 30 J/m2 and kept for 4 h in the presence of EdU and HU (1BR.3 fibroblasts; left panel) or were irradiated and kept for 4 h in the presence of HU and in the presence/absence of EdU (VH10 fibroblasts; right panel). Next, cells were subjected to aniFOUND and the isolated material was used for western blot. Material from 1 million cells was loaded for inputs while for the pull-down the whole amount was loaded. The membrane was labelled with the indicated antibodies using two discriminable secondary antibodies with different wavelengths.