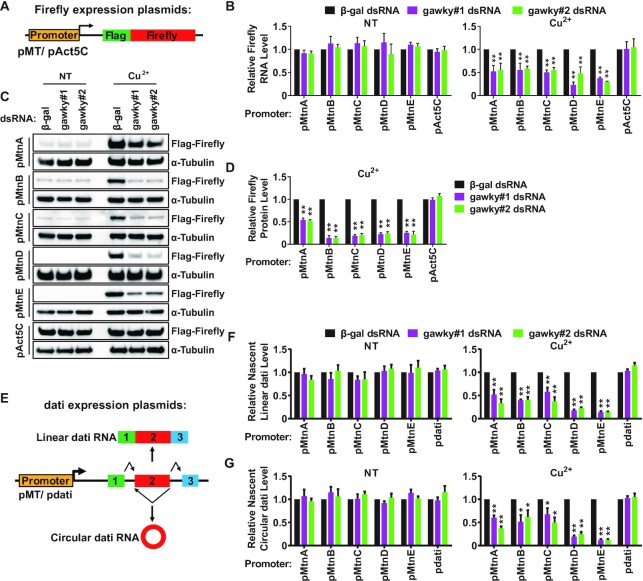
Figure 4.
The regulatory effect of gawky on transcription depends on the promoter. (A) Schematic representation of FLAG-tagged firefly luciferase expression plasmids. The MT promoters or the Act5C promoter was inserted upstream of the transcriptional start site. Stable cell lines were generated using these plasmids. (B–D) S2 cell lines stably expressing FLAG-tagged firefly luciferase were treated with two independent gawky dsRNAs for 3 days followed by the addition of 500 μM copper for the final 12 h to induce metal stress. (B) The expression of firefly luciferase mRNA was subjected to real-time RT-PCR analyses following RNA isolation. Data were normalized to the β-gal dsRNA sample and are shown as mean ± SEM. n = 3. ∗∗P < 0.01; ∗P < 0.05. (C) Firefly luciferase protein was subjected to western blotting analyses using equal amounts of protein extracts from whole cells. α-Tubulin served as a loading control. Representative blots are shown. n = 3. (D) The protein level of the copper-induced firefly luciferase in (C) was quantified using ImageJ from three independent western blotting experiments. To provide better representation of the levels of non-induced proteins, we increased the amount of protein used for western blotting analyses (see Supplementary Figure S6C and D). Data were normalized to the β-gal dsRNA sample and shown as mean ± SEM. ∗∗P < 0.01; ∗P < 0.05. n = 3. (E) Schematic representation of dati expression plasmids that can generate linear RNAs (exon 1, exon 2, and exon 3 are joined in a linear order) and circular RNAs (the end of exon 2 is joined to the beginning of exon 2). The MT promoters or the dati promoter was inserted upstream of the transcriptional start site. (F, G) After gawky RNAi, S2 cells were transfected with different dati expression plasmids for 2 days followed by the addition of 500 μM copper for the final 12 h to induce metal stress. The level of nascent linear (F) or circular (G) dati RNA was measured by real-time RT-PCR using RNA extracts from nuclear run-on experiments. Data were normalized to the β-gal dsRNA sample and are shown as mean ± SEM. n = 3. ∗∗P < 0.01; ∗P < 0.05.