Abstract
Lake Malawi cichlid fishes exhibit extensive divergence in form and function built from a relatively small number of genetic changes. We compared the genomes of rock- and sand-dwelling species and asked which genetic variants differed among the groups. We found that 96% of differentiated variants reside in non-coding sequence but these non-coding diverged variants are evolutionarily conserved. Genome regions near differentiated variants are enriched for craniofacial, neural and behavioral categories. Following leads from genome sequence, we used rock- vs. sand-species and their hybrids to (i) delineate the push–pull roles of BMP signaling and irx1b in the specification of forebrain territories during gastrulation and (ii) reveal striking context-dependent brain gene expression during adult social behavior. Our results demonstrate how divergent genome sequences can predict differences in key evolutionary traits. We highlight the promise of evolutionary reverse genetics—the inference of phenotypic divergence from unbiased genome sequencing and then empirical validation in natural populations.
Subject terms: Developmental biology, Evolution, Neuroscience
Introduction
Our understanding of how the genome encodes natural variation in form and function is still limited. This is the case for almost any trait, from height to behavior to complex disease1. The reasons for this are manifold, but they include an underappreciated role of non-coding genetic variants linked to differences in traits. This is apparent in our assumptions and in syntheses of data. For instance, only 25 years ago, experts thought that the human genome might contain 70,000 to over 100,000 genes to account for our complexity2. More recently, it has been estimated that upwards of 93% of human disease related variants—traits for which we have the most data from genome wide association studies (GWAS)—reside in noncoding DNA sequence3. Many of these noncoding variants are regulatory, that is, they affect the expression of genes4. Moreover, gene expression differences are likely to play a significant role in the evolution of closely related species5,6. Therefore, despite a refined understanding of how single genes work in controlled cellular environments, it remains unclear how the genome is activated to produce natural phenotypes, and this may be particularly vexing for context-dependent processes like development or behavior.
Over the past two decades, systems have been developed to identify the genetic basis of traits from nature7. Amongst vertebrate animals, these traits include body armor8, color9, head and jaw shape10–12, parental care13,14, song15 and coordinated movement16. The take home message from this work has been that a small number of genes from recognizable pathways explain a considerable proportion of phenotypic variance. Yet, these studies may be biased in interpretation and limited in inference space. The focus is typically on one or two species and one trait at a time, often using hybrid pedigrees founded by a small number of individuals, and candidate gene or QTL approaches. Here, we explored a different strategy in a system of many species with many divergent traits. We sought to determine the genetic differences between closely related groups of species and then to focus experiments on leads from genome divergence. In essence, we’ve asked the genome which traits to follow. This approach is similar to genome-first examples in mammals17,18 wherein unbiased analysis of genome data selects the traits rather than a trait-first model like QTL mapping.
The Malawi cichlid system is an apposite one for our research aims. The assemblage comprises hundreds of closely related species that have diversified in the last 500,000 to one million years19, such that the genomes of individuals across species boundaries remain highly similar20. An appreciable fraction of genetic polymorphism identified in Malawi species is shared with cichlid lineages from throughout East Africa—suggesting that ancient genetic variation fuels diversification of the Malawi flock21. Set against this background of genome similarity, Malawi cichlids exhibit staggering diversity in phenotypes including pigmentation22, sex determination23,24, craniofacial and brain patterning10,25,26 and social behavior27–29. Previous work has focused on the genomic and early developmental underpinnings of this diversity, in rock- vs. sand-dwelling species20,25,26,30.
Rock- vs. sand-species form ecologically distinct groups similar to other ecotypes in well-known evolutionary radiations (marine vs. freshwater sticklebacks; tree vs. ground finches and anoles)31. The main difference in this case is that each of the rock- and sand- groups contains more than 200 species. Recent divergence, rapid speciation and meta-population dynamics synergistically lead to the broad sharing of polymorphism across the rock-sand speciation continuum21,32. Malawi rock-dwellers tend to be strongly territorial and aggressive; they breed and feed at high density in complex rock-reef habitats. Most eat algae from the substratum with strongly reinforced jaws packed with teeth. Adult rock-dweller brains exhibit enlarged anterior components, telencephala and olfactory bulbs. Sand-dwellers are less site-specific and less aggressive. They often breed on communal leks where males build sand ‘bowers’ to attract females33. Many capture small prey using acute vision and fast-moving gracile jaws; their brains and sensory apparatus are elaborated for more posterior structures optic tecta, thalamus and eyes (Supplementary Fig. S1). We aimed to understand evolutionary divergence between rock- and sand-dwelling lineages by identifying the number, type and spectrum of genetic variants that separate these groups.
To target this primary axis of evolutionary divergence in the Lake Malawi species assemblage31, we compared whole genomes of one male individual each from 8 rock-dwelling and from 14 sand-dwelling species (Supplementary Table S1), to an average of 25X coverage per individual. Species were chosen to represent the diversity present within each of the rock- and sand-groups (Fig. 1A), in terms of body size, color pattern, ecology and phylogenetically defined lineages within the sand- species group32.
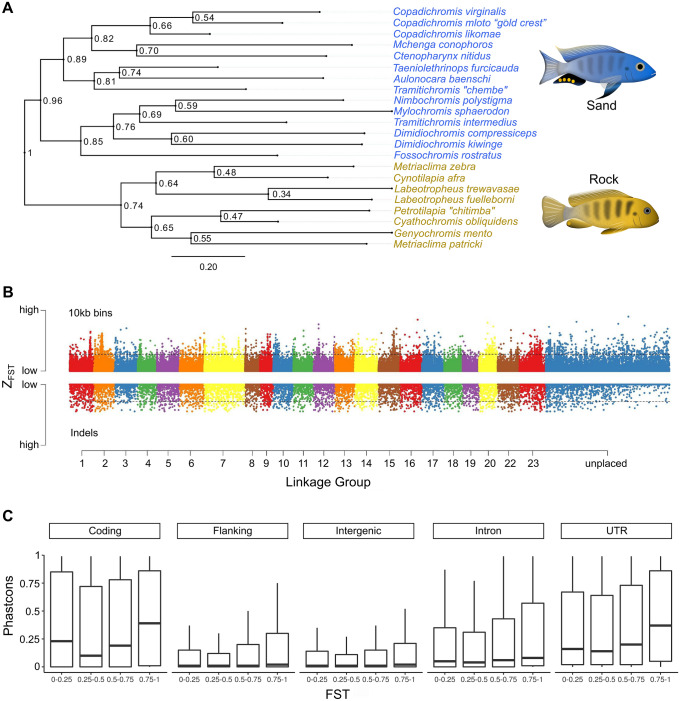
Figure 1.
The genomic substrate for rock vs. sand evolution. (A) A maximum likelihood phylogeny of eight rock- and 14 sand-dwelling species, based on variable sites (informative SNP and InDels) identified throughout the genome. “Sand” species contain representatives of the lineages “shallow benthic, deep benthic and utaka” from Ref.32, while the “rock” species correspond to the “mbuna” lineage. (B) A plot of Z-FST (FST normalized using Fisher’s Z-transformation) across the genome (ggplot2 v3.3.3https://ggplot2.tidyverse.org/), plotting genomic divergence between rock- vs. sand-dwelling groups. Single nucleotide polymorphisms (SNPs) summed over 10 kb bins and insertion-deletion mutations (InDels) are shown on the same scale. Numbers along the x-axis refer to linkage groups (i.e., chromosomes) and threshold lines indicate 2.5% FDR. (C) Evolutionary conservation (PhastCons) scores were calculated for each nucleotide across the genome, subdivided by genome annotation and plotted by bins of increasing FST. The PhastCons score for each genome category is significantly higher for increasing bins of FST (pairwise Wilcoxon rank sum p value < 2e−16 for each pair of bins).
Results
The genomic signature of rock-sand divergence
We compared the genomes of 8 rock dwellers and 14 sand dwellers to uncover the genomic signature of rock- versus sand-evolutionary diversification. We aligned sequence data to a reference genome of nearly 1 gigabase34 and identified approximately 22 million Single Nucleotide Polymorphisms (SNPs) and 200,000 Insertion-Deletions (InDels). We calculated FST per variant, and averaged across 10 kb windows, to quantify divergence between rock and sand species. We found that 0.06% of SNPs and 0.44% of InDels are alternately fixed between rock- and sand-groups. When these divergent variants and genome regions (2.5% FDR) are mapped to linkage groups (chromosomes), it is apparent that the signature of rock- vs. sand-divergence is distributed relatively evenly across the chromosomes (Fig. 1B). Among fixed variants, 3.5% were found in coding regions and 96.5% were predicted to be non-coding; ~ 17% in intergenic regions, 38% in introns, 38% in flanking regions (within 25 kb up- or down-stream of a gene), and 3% in annotated UTRs. Rock vs. sand fixed coding variants were more likely to be missense/loss-of-function (72.6%) than silent (27.3%).
We next generated whole-genome alignments of five published cichlid reference genomes from across East Africa35 and estimated an evolutionary conservation score for each nucleotide position. Akin to phylogenetic footprinting, this approach allows inference of function for regions that are slower to change than others due to the long-term effect of purifying selection. For both coding and non-coding portions of the genome, we found that rock-sand divergence correlates positively with evolutionary conservation scores (Fig. 1C), suggesting that differentiated rock-sand variants, including many non-coding variants, are enriched for function.
4484 genes lie within 25 kb of either an alternately fixed variant or a highly divergent 10 kb window (2.5% FDR). Pathway enrichment analysis36 of human homologs/analogs for these genes reveals categories spanning early embryonic development, craniofacial morphogenesis, brain development, synaptic transmission and neuronal function (Supplementary Table S2). In particular, rock-sand divergent genes are enriched for GO Biological Process terms ‘telencephalon development’ (p < 1.7e−18), ‘adult behavior’ (p < 2e−14), ‘synaptic plasticity’ (p < 1.4e−12), ‘odontogenesis’ (p < 3.7e−11), ‘response to BMP’ (p < 3.2e−09), ‘gastrulation’ (p < 5.6e−06), ‘face morphogenesis’ (p < 8.9e−08), ‘neural crest cell differentiation’ (p < 4.3e−13), and ‘eye development’ (p < 1.3e−15). Over-represented gene families included nuclear hormone receptors (p < 3.0e−08), HOXL subclass homeoboxes (p < 1.4e−07), TALE class homeoboxes (p < 4.2e−04) and Forkhead boxes (p < 9.33e−04; for novel expression domains in cichlid foxp2 see Supplementary Fig. S2). We observed enrichment for the mouse phenotypes ‘abnormal cognition’ (p < 3.3e−15), ‘abnormal learning and memory’ (p < 2.6e−15), ‘abnormal craniofacial morphology’ (p < 4.9e−11) and ‘abnormal social/conspecific interaction’ (p < 7.1e−14). We used the list of differentiated genes to query an Allen Brain Atlas dataset that reports gene expression in hundreds of brain regions37. Rock- sand-divergent genes were enriched for the basomedial nucleus of the amygdala, a sub-region of the telencephalon (padj = 0.001) that regulates fear, anxiety, and physiological responses to territorial intruders in rodents38,39, and has been linked to Social Anxiety Disorder in humans40. Finally, we queried databases of genes involved in human disease. Genes near divergent variants are significantly enriched for factors implicated in neurological disease like Autism Spectrum Disorder (SFARI41, Fisher’s exact test p value < 2e−16) and disorders related to the neural crest42, (Fisher’s exact test p value < 2e−16).
Given the prevalence of evolutionarily conserved, non-coding, divergent rock-sand variants and genome-wide enrichment for craniofacial and neural crest biology, we examined overlap with published datasets of mammalian neural crest and craniofacial enhancers43,44. These data allow us to identify craniofacial and cranial neural crest cell (CNCC) enhancers conserved between mammals and cichlids and fixed variants between rock and sand species within these conserved regulatory elements. A total of 275 craniofacial enhancer elements and 234 human CNCC enhancers are evolutionarily conserved between mammals and cichlids. We found divergent rock-sand mutations within the enhancer elements of key genes integral to CNCC specification and migration (Supplementary Table S2). Notably, from both datasets, fixed rock-sand variants were found within the enhancer region of the gene nr2f2, a nuclear receptor and master neural crest regulator45. Rock-sand divergent variants were similarly located within craniofacial enhancers of three genes (yap1, fat4, rere) that function in the Hippo signaling pathway, as well as within enhancers of irx3 and axin2. These data linking rock-sand fixed SNP/InDels to evolutionarily conserved, experimentally verified enhancers further underscore the importance of non-coding variation in the craniofacial evolution of rock- and sand-lineages46.
Genome-wide divergence between rock vs. sand Malawi cichlids involves a relatively small percentage of genetic variants. Divergent variants are (a) predominantly non-coding, (b) in long-term evolutionarily conserved loci (c) enriched for genes and pathways involved in embryonic development, brain development, brain function and behavior, and craniofacial morphogenesis. Given these strong patterns of enrichment, we used the experimental power of the Malawi cichlid system to interrogate features of early development and adult behavior that differ between rock- and sand-groups.
A gastrula-stage map of forebrain diversification
Rock- vs. sand-dwelling Malawi cichlids exhibit divergence in or near genes enriched for BMP signalling, gastrulation, eye and telencephalon development, as well as the TALE (Irx) gene family. To explore the developmental consequences of this differentiation, we investigated early forebrain specification in rock- and sand-embryos, building upon our previous studies and interest in irx1b and early brain development25,26. During development, the complexity of the vertebrate brain is first laid out in the neural plate, a single-cell thick sheet of cells that forms between non-neural ectoderm and the germ ring at gastrulation. Irx genes act as transcriptional repressors of BMP signal in gastrulation, and function to specify the neural plate47. BMPs, in turn, are protective of the anterior-most region of the neural plate, which will ultimately give rise to the telencephalon, and suppress the eye field48. Given alternatively fixed variants in the irx1b gene, expected interactions between Irx and BMP signaling in the early embryo and known telencephalon vs. eye size differences between rock- vs. sand-species25,26, we examined and quantified the early activity of irx1b and BMP in rock- vs. sand-embryos.
We used a custom device to orient and image cichlid embryos in toto at gastrula and neurula stages49. In early gastrula (EG), irx1b (red) and BMP signal (green, PSMAD) delineate complementary dorsal and ventral domains of the embryo (Fig. 2A). By mid-gastrula (MG), irx1b shows two expression domains, one in the posterior portion of the developing neural plate (np) and the second co-expressed with PSMAD activity around its anterior border (white arrowheads). By late gastrula (LG), the domains of irx1b expression and PSMAD activity sharpen around the leading edge of the neural plate but remain overlapping around the periphery. Notably, irx1b expression is expanded in the anterior domain of sand-dwellers (S) compared to rock-dwellers at EG and MG, and then defines the boundary of the neural plate earlier in sand-dwellers (S) in LG (arrowheads). As a consequence, BMP signal should have a longer-lasting influence on the neural plate in rock-dwelling species. Based upon manipulative experiments in zebrafish48, this is predicted to result in a relatively larger presumptive telencephalon and smaller eye field.
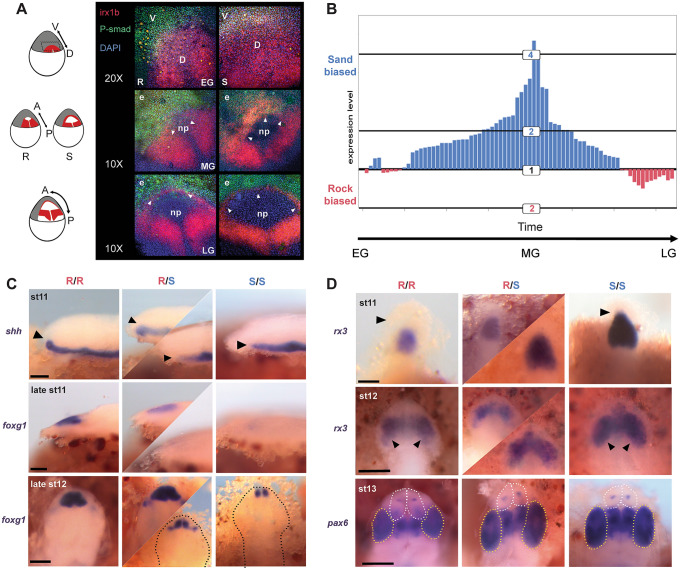
Figure 2.
A gastrula-stage map of forebrain diversification. (A) Double in situ hybridization—immunohistochemistry to visualize irx1b expression (red) and BMP [PSMAD] activity (green) across the stages of cichlid gastrulation; DAPI in blue. Three rows represent early, mid and late gastrulation (EG, MG, LG) in embryos of rock- (R) and sand-dwelling (S) species. Sand-dwellers show expanded irx1 expression in the dorsal portion of the embryo at EG, expanded irx1b expression in the anterior domain at MG (arrowheads) and clear PSMAD activity from the developing neural plate (np) earlier in LG (arrowheads). e epidermis. Schematics at left show irx1b expression domains in red, on cartoons of cichlid embryos. (B) Relative expression of rock (red) and sand (blue) irx1b alleles, sampled from 74 heterozygous rock-X sand-F2 embryos, across the stages of gastrulation. F2 embryos were sampled at stage 9 (gastrulation). Because Malawi cichlid species are maternal mouthbrooders and eggs are fertilized in batches per brood, each brood’s embryos vary in timing of fertilization by up to 4 h. Embryos within broods can therefore be sub-staged in gastrulation (see “Materials and methods” section). Each bar on the plot represents the relative allelic expression of sand- and rock-irx1b in a heterozygous F2 individual. Quantification of allele-specific expression (ASE) shows that levels are sand-biased, and that this effect is strongest in MG. (C) In situ hybridization of shh and foxg1, during neurula stages, showing development of the telencephalon in rock-X sand-F2 embryos, indexed for irx1b genotype. F2 individuals homozygous for rock-irx1b alleles (R/R) show a more dorsal progression of shh expression (black arrowheads), an earlier and a larger expression domain of the telencephalon marker foxg1. The top two rows are lateral views; bottom row is a dorsal view. Dotted lines demarcate the outline of the embryo in dorsal view. Heterozygous individuals exhibit greater variation in expression domains (middle columns), indicating that genetic factors other than variants in irx1b contribute to this phenotype. (D) In situ hybridization for rx3 and pax6, during neurula stages, chart the development of the eye field in rock- X sand- F2 embryos indexed for irx1b genotypes. F2 individuals homozygous for sand-irx1b alleles (S/S) show larger domains of rx3 (black arrowheads) and larger eyes (pax6, also marked by yellow dotted line), but smaller telencephala (white dotted line). All panels are dorsal views. Heterozygous individuals exhibit greater variation in expression domains (middle columns), indicating that genetic factors other than variants in irx1b contribute to this phenotype.
We developed a panel of rock-x sand-hybrid crosses to formally evaluate the role of irx1b in forebrain diversification. First, we used quantitative RT-PCR to measure allele-specific expression (ASE) in heterozygous rock-x sand-F2 hybrids, across the stages of gastrulation. We observed that the sand-irx1b allele was expressed at significantly higher levels (average of 2.5-fold; p = 4.5e−13; Student’s t-test) and that this difference was largely confined to MG (Fig. 2B). Next, we used hybrid embryos to chart the development of the telencephalon and the eye field. Rock-x sand-F2 hybrids, indexed for irx1b genotype, were raised to neurula and somitogenesis stages and we examined the expression of shh (which induces foxg1 and the ventral forebrain), foxg1 (a marker of the telencephalon), and rx3 (a marker of the eye field), by in situ hybridization. F2 individuals homozygous for rock-irx1b alleles exhibited a larger and more rostral domain of shh expression, an earlier and larger domain of foxg1 and a smaller rx3 domain (Fig. 2C,D). These differences between rock- vs. sand-irx1b genotypes match expression divergence observed amongst rock- vs. sand-species25,26. Finally, when we compared the relative size of the telencephalon among irx1b genotypes, individuals homozygous for rock-alleles exhibited larger telencephala (p < 0.0001, Tukey’s test, Supplementary Fig. S3). We conclude that genetic variants in and around the irx1b gene contribute to divergent specification of the Malawi cichlid forebrain, likely via spatial, temporal and quantitative variation in the expression of irx1b itself.
Our genome sequencing revealed a near-fixed InDel in the 3′ UTR of the Malawi cichlid irx1b gene (Supplementary Fig. S4). Rock-species possess an 85 bp insertion, compared to cichlid species from outside of the Malawi lineage. Sand-dwellers largely lack the insertion and exhibit a 6 bp deletion compared to outgroups. The insertion shows strong genetic similarity to a fragment from the Rex1 family of non-LTR retrotransposons50. Given the likelihood that Astatotilapia calliptera populations surrounding Lake Malawi may have seeded the Malawi evolutionary radiation and contributed to rock- and sand-dwelling lineages21,32, we explored the presence/absence of this InDel in Astatotilapia samples. We found that most Astatotilapia individuals and populations had the rock-irx1b allele (the insertion), but that an individual from Chizumulu Island was fixed for the sand- allele and two individuals sampled from Itupi were heterozygous. Because the 85 bp insertion in rock-species is a partial Rex1 fragment, and sand- species carry a 6 bp deletion compared to outgroups, we speculate that the current rock- and sand-divergent alleles were generated by at least two imperfect excision events of an element that invaded the genome of the Malawi + Astatotilapia ancestor. Rex1/Babar retrotransposons have been active in African cichlid genomes, and are known to influence gene expression when inserted in 5′ and 3′ UTRs35. Future experiments will determine whether this Rex1 insertion causes the differences in irx1b gene expression and forebrain specification we observed.
Genomics of divergent social challenge and opportunity
Rock- and sand-dwelling Malawi cichlids live in strikingly different social and physical environments. Rock-dwelling males tend to be more aggressive than sand-dwellers27 and defend territories year-round as sites for feeding and breeding. By contrast, sand-dwellers are more exploratory than rock-dwellers29 and only breeding males tend to be territorial, often building sand bowers to attract females and mitigate male–male aggression. Given these observations and genome-wide enrichment for categories related to adult behavior and social interaction, we designed an experimental paradigm to investigate brain gene expression profiles associated with divergent rock- vs. sand-social behaviors.
We evaluated social challenge and opportunity amongst males using a large tank with a ‘rock’ habitat at one end and ‘sand’ at the other, separated by glass bottom (Fig. 3A). When parental rock-species are placed in this tank paradigm, males court females on the rock side of the tank. Males of sand-species court females over sand and construct species-appropriate bowers. When single hybrid rock-x sand-F1 males are placed in this arena with hybrid F1 females, males invariably court females over the ‘rock’ habitat. However, when two rock-x sand-hybrid F1 males (brothers) were allowed to compete for gravid hybrid F1 females in this tank paradigm, we observed something different. One male, typically the larger, courted females over the rock habitat, and the other simultaneously constructed bowers to court females over the sand. We found no difference in gonadal-somatic index (GSI), an established biological metric of reproductive status and maturity51, between F1 males behaving as ‘socially rock’ vs. ‘socially sand.’ (Supplementary Fig. S5). Our observation of divergent behavior between F1 brothers in the same tank suggests an interaction between the genome and the social environment.
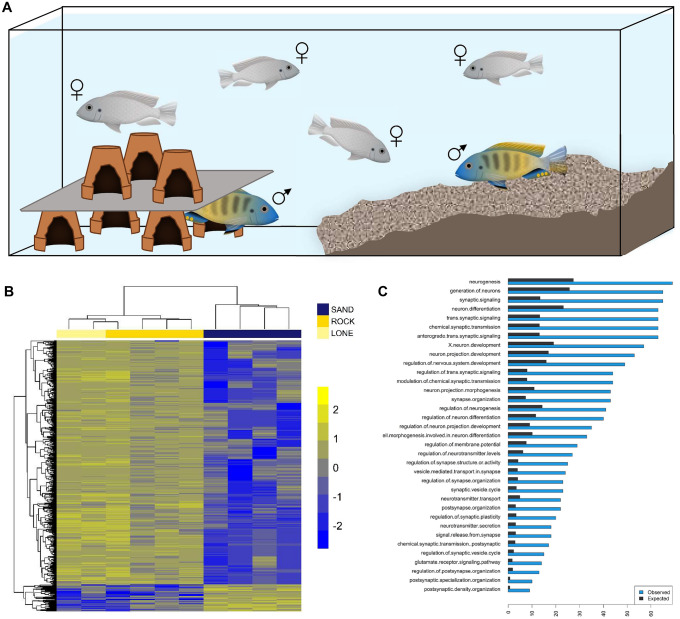
Figure 3.
Genomics of divergent social context. (A) Schematic of the social context behavioral paradigm, in which rock-, sand- and rock-X sand-F1 hybrids were evaluated. (B) Heatmap of genes (ggplot2 v3.3.3 https://ggplot2.tidyverse.org/) differentially expressed in the brains of F1 males behaving in either rock or sand social contexts. Each row in the heatmap is a gene, each column is an individual. R individuals are socially rock, S individuals are socially sand and the numbers indicate sibling pairs. Two F1 males were not paired with other males, and courted females over the rock habitat (lone). All other F1 males (n = 8) were introduced to the testing arena in dyads. Notably, male brain gene expression clusters by social context and not fraternal relationships. (C) Gene Ontology (GO) Biological Process terms that show greater than expected overlap between (i) genes differentially expressed in the brains of social rock- vs. social sand-males and (ii) genes that are differentiated in the genomes of rock- vs sand-species groups.
We used RNA-seq to investigate gene expression profiles associated with behavior of rock-x sand-F1 hybrid males that were actively courting females over rock vs. sand. Whole brains of F1 males tested singly (n = 2 lone) as well as F1 brothers assayed in dyads (n = 4 dyads) were collected during courtship, and interrogated by RNA-seq. Strikingly, gene expression profiles clustered not by fraternal relatedness, but rather by behavioral context (Fig. 3B). Males from dyads that courted females over rocks had expression profiles similar to single males (who also courted over rocks) but distinct from their brothers that built bowers and courted females over sand in the same tank. Genes were considered significantly differentially expressed between ‘social rock’ and ‘social sand’ brains if they exhibited both a fold change ≥ 2 and crossed the threshold of padj < 0.05. Based on this criterion, we found 832 genes differentially expressed between rock- vs. sand-behaving males (Fig. 3B, Supplementary Table S3). Among differentially expressed genes, we observed significant functional enrichment for GO Biological Process categories ‘synaptic signaling’ (p < 2.3e−21), ‘synaptic plasticity’ (p < 3.6e−09), ‘visual behavior’ (p < 2.09e−06); mouse phenotypes ‘abnormal learning/memory/conditioning’ (p < 5.9e−07), ‘abnormal telencephalon morphology’ (p < 3.95e−07), ‘abnormal spatial learning’ (p < 9.9e−07) and pathways ‘axon guidance’ (p < 3.3e−05), ‘oxytocin signaling’ (p < 5.2e−05) and ‘estrogen signaling’ (p < 1.8e−04) (Supplementary Table S3). Matches against the Allen Brain Atlas database of gene expression yielded enrichment for sub-regions of the telencephalon: CA3, hippocampus (padj = 4.6e−6), CA2, hippocampus (padj = 7.3e−5), CA4, hippocampus (padj = 0.001), claustrum (padj = 0.001), subiculum (padj = 0.003), dentate gyrus (padj = 0.03) and the basomedial nucleus of the amygdala (padj = 0.03). The hippocampus encodes episodic memory and spatial representations of the environment52, and more recently its subregions have been shown to play critical roles in anxiety, social interaction, and social memory formation53–55. Roughly 38% of differentially expressed genes also contained genetically differentiated SNP/InDels between rock- and sand- species (p-value < 2e−6, Fisher’s exact test), implying considerable cis-acting genetic variation. Enrichment of numerous categories related to brain function and synaptic plasticity showed greater overlap than expected (Fig. 3C). These context-dependent differences suggest concerted changes in brain gene expression as males experienced and responded to different social challenges and opportunities28,56.
Discussion
A fundamental problem in evolutionary biology is understanding the cellular, developmental and genetic basis of how traits change. This is a challenge because we lack sufficient information about how genes work in outbred genomes from nature and we do not fully comprehend the causal role of noncoding variation in specifying form and function. This problem is especially difficult for traits that are only observed in particular contexts, like development and behaviour. To make progress, we and others have focused on study systems exhibiting abundant phenotypic diversity built from a relatively small number of genetic changes. Here we identify and characterize the genetic variants that demarcate one of the deepest evolutionary splits amongst Lake Malawi cichlid groups, that between rock- and sand-dwelling species thought to have diverged in the last one million years. We found a small percentage (less than 0.1%) of genetic variants to be differentiated between rock- and sand-groups, and that the majority of differentiated variants (> 96%) were noncoding. Differentiated non-coding variants were more likely to be in an evolutionarily conserved locus as a function of genetic differentiation, suggesting that divergent rock- vs. sand-noncoding changes are functional. To support this idea, we identified alternately fixed rock- vs. sand-noncoding variants within experimentally verified, vertebrate-conserved craniofacial and cranial neural crest cell enhancers. The latter observation is similar in type to the discovery of human-specific deletions within mammal-conserved regulatory sequence18.
Recently we surveyed genome-wide divergence between sand-dweller sub-groups that construct pit vs. castle bowers, sand-made structures to attract females for mating28 . Mapping those variants to the same genome reference, we expected distinct patterns of diversification because rock-sand and pit-castle divergence likely occurred at different times, along different trait axes, under the control of different evolutionary forces31. Consistent with expectation, there is clear clustering of genome divergence on chromosome 11 for the pit-castle comparison (Supplementary Fig. S6), while all chromosomes carry the signature of rock-sand diversification (Fig. 1B). However, contrary to our expectation, rock- vs. sand- and pit- vs. castle-radiations have diverged in similar gene sets. Out of 3070 genes identified near 10 kb high FST regions in the rock- vs. sand-comparison, 483 overlap with 1090 genes identified near high FST regions in the pit- vs. castle-comparison (p-value < 2e−9, Fisher’s exact test, Supplementary Table S4). This result may imply that evolutionary diversification in Lake Malawi is limited, or constrained, by chromosomal location.
Overall, genes in proximity to rock-sand divergent variants were enriched for functional categories related to early forebrain and craniofacial development, neuronal function and social behavior. This list of variants, coupled with consistent patterns of functional and pathway enrichment, motivated follow up experiments focused on early brain development and adult social behavior (Fig. 4). It is apparent from our work here and previously25,26, that Malawi cichlid brains and nervous systems begin to differ during gastrulation in pathways that can be predicted from divergent genome sequences. This is interesting for at least two reasons. First, this observation runs counter to the ‘late equals large’ textbook example57 of how brains evolve differences in relative proportions of their parts26, and along with other reports58 suggests that variation in early brain development may be a significant factor in brain evolution. Overall, such early variation in development is not thought to be a driving force in evolution, precisely because early changes can have global and ramifying effects. Collectively, our findings provide a partial description of the conditions wherein variation during the earliest stages of development can contribute to evolutionary diversification. In each case we have examined, variation in gene expression is quantitative, heterochronic and limited to a precise stage or time period.
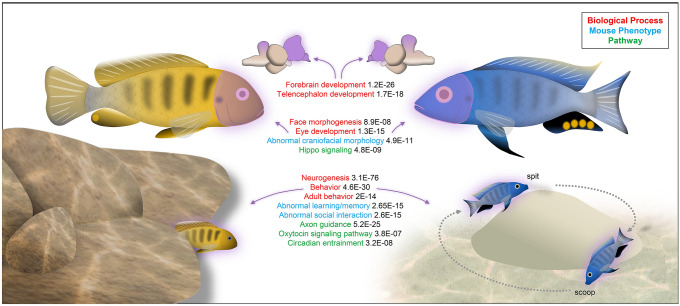
Figure 4.
Genome-enabled discovery of evolutionary change in morphology and behavior. Summary cartoon synthesizing significant enrichment categories that differentiate the genomes of rock- vs. sand-dwelling Malawi cichlids. Strong and consistent enrichment of craniofacial, neural and behavioral categories motivated follow-on experiments in early brain development (Fig. 2) and adult social behavior (Fig. 3).
Sydney Brenner recognized the relationship between the genetic specification of nervous systems and the behavioral output of the brain59. However, because these events take place so far apart in the lifespan of a vertebrate, they are rarely studied simultaneously. Here, the genome connects the two phenomena: rock- vs. sand-divergent gene sets and strong functional enrichment indicate that both brain development and social behavior have contributed to the evolutionary diversification of these groups. To evaluate social behavior in rock- vs. sand-dwelling Malawi cichlids, we constructed a social context arena. The presence of sand and simulated rocky caves was sufficient to elicit species-appropriate male behavior when rock- or sand-males were tested with rock- or sand-gravid females. When rock-x sand-F1 hybrid males were tested, one per tank, with F1 hybrid females, males courted females in the rock quadrant of the tank. Notably, when dyads of F1 brothers were tested in this tank paradigm with gravid F1 females, we observed simultaneous ‘social rock’ and ‘social sand’ behavior. Brain gene expression profiles from behaving males clustered by social context (social rock vs social sand), and not by fraternal relationships. Differentially expressed genes were enriched for brain regions and pathways implicated in social interaction and overlapped significantly with rock- vs. sand-divergent genetic variants.
Social context is known to influence the brain. For instance, our clustering results are similar to those of Whitfield and colleagues (Whitfield et al. 2003) who showed that brain gene expression in honey bees was predictive of behavior. Likewise, changes in brain morphology and gene expression predictably accompany the ascent to dominance in the cichlid fish Astatotilapia burtoni60. Our data seem not to fit the model of dominant-subordinate however. In our experiments, the gonado-somatic index (GSI) did not differ between social rock vs. social sand brothers within dyads. Both males exhibited nuptial coloration, courted females and in cases with multiple gravid females, both brothers reproduced. Body size was associated with divergent social rock vs. social sand behavior of F1 males; the social rock brother was always larger (mean mass was 26.96 g ± 3.4 [SE] compared to 19.45 g ± 2.2).
Our experiments demonstrate that F1 hybrid male brains can express both social rock- and social sand-behavioral programs, and that social context determines which program is executed. This observation is similar to, but also different than, pit-digging × castle-building F1 male Malawi cichlids who carry out parental bower behaviors in a specific sequence28. Notably, in both cases, the hybrid males exhibit one of the two different parental behaviors at any one time—there is no intermediate behavior. In the pit- vs. castle-case, we think that the bower structure itself and/or a threshold signal from females might lock the hybrid male brain into a behavioral state. In the rock- vs sand-case here, it appears that other social cues (i.e., the presence and size of a rival male) lock the hybrid male brain into a behavioral state. These context-dependent behaviors, accompanied by changes in brain gene expression, are compelling examples of interaction between the genome and the social environment. The cellular and genetic basis of these behaviors and their plasticity deserves further attention. Here, our comparative genomic and brain gene expression data, combined with enrichment testing and experimental approaches, highlight that the Malawi cichlid telencephalon will be central to this work. The telencephalon is similarly likely to be important for other Malawi cichlid social behaviors, including aggression and bower building27,28. Overall, our data add to the reappraisal of the telencephalon as an anatomical nexus of evolutionary innovation across vertebrate lineages61.
Materials and methods
Genome sequencing
We extracted genomic DNA, from fin clips of 22 male individuals62 (Qiagen DNeasy, Cat #69504), from 8 rock dwelling and 14 sand dwelling Lake Malawi species (Supplementary Table S2) representing broad diversity across the rock and sand lineages in Lake Malawi. We made libraries using the Illumina Nextera Library prep kit and performed paired-end sequencing on the Illumina Hi-Seq 2500 at Georgia Tech. The Metriaclima zebra reference genome version MZ_UMD2a34 was used for genome alignment, variant discovery and annotation using standard BWA63 and GATK practices64. The maximum likelihood tree in Fig. 1A was constructed using SNPhylo65, from variant data.
Genetically divergent regions
Vcftools66 was used to calculate FST (–weir-fst-pop) between the 8 rock and 14 sand species excluding SNPs with less than half variants called across species (–max-missing 0.5). Variants with FST = 1 were noted to be alternately fixed between rock and sand lineages in our dataset. FST was also measured across 10 kb windows (–fst-window-size). Significance thresholds were marked using the fdrtool67 package in R. All variants were annotated using Snpeff 4.3i68. We tested the genes within 25 kb of significantly differentiated variants for enrichment of functional categories. The cichlid gene names were converted to human analogs using Treefam based mapping69 and functional enrichment was determined using the TOPPFUN web-browser interface70.
PhastCons analysis
Pairwise alignments were generated using lastz v1.0271, with the following parameters: “B = 2 C = 0 E = 150 H = 0 K = 4500 L = 3000 M = 254 O = 600 Q = human_chimp.v2.q T = 2 Y = 15,000”. This was followed by using USCS genome utilities (https://genome.ucsc.edu/util.html, https://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/FOOTER) axtChain tool with -minScore = 5000. Additional tools with default parameters were then used following the UCSC whole-genome alignment paradigm (http://genomewiki.ucsc.edu/index.php/Whole_genome_alignment_howto) in order to obtain a contiguous pairwise alignment. Multiple alignments were generated from pairwise alignments with the multiz v11.272 program, using default parameters and the following pre-determined phylogenetic tree: ((((M. zebra, P. nyererei), A. burtoni), N. brichardi), O. niloticus) in agreement with Brawand et al.35. Sequence conservation scores were then obtained using PhastCons73 with a phylogenetic model estimated by the phyloFit74 program, both from the PHAST software package (v.1.3). The model fitting was done using default parameters. PhastCons was run in two iterations, first to obtain the free parameters of the model (-estimate-trees and -no-post-probs) and then using the output from this we ran PhastCons again to attain the conservation scores with—target-coverage 0.3—expected-length 100.
Vertebrate-conserved enhancer elements
A comparative genomic approach was used to identify putative craniofacial and neural crest CNEs in mammals that segregate SNPs between rock-sand cichlid species. Experimentally verified and published genome-wide craniofacial and neural crest enhancers active during early embryonic stages that play a role in shaping the development of neural crest and craniofacial structures in mammals were identified from published literature43,44. We used the liftOver tool75, which maps orthologous genomic regions between species to convert genomic coordinates from one species to another. Using a Human to Oreochromis niloticus to Metriaclima zebra mapping and a Mouse to Oreochromis niloticus to Metriaclima zebra mapping, we identified the orthologous genomic locations of the published craniofacial and neural crest enhancers in cichlids. We designated any alternately fixed variant (variant with FST = 1) that was also within an orthologous CNE as putatively involved in the rock-sand divergence (Supplementary Table S2).
Brain region enrichment analysis
We identified 10,391 cichlid genes with human homologues and generated an expression matrix for each gene across 250 human brain structures spanning telencephalon, diencephalon, mesencephalon, and metencephalon using adult human brain microarray data collected by the Allen Brain Institute37. Cortical regions and gyri for which fish do not have putative homologues were excluded from the analysis (100/350, leaving 250 regions for subsequent analysis). The expression matrix was generated using the get_expression function in the ABAEnrichment Bioconductor package in R76. We then calculated the specificity of expression for each gene in each of these brain regions using the specificity.index function in the pSI package for R. This function calculates a matrix of gene expression specificity indices, and corresponding p-values, as described previously77,78. We then tested whether (1) genes within 25 kb of rock vs. sand significantly differentiated variants (described above under “Genetically Divergent Regions”), and (2) genes that were differentially expressed between rock- vs. sand-behaving F1 hybrid males, were enriched for transcriptional markers of specific brain regions using the fisher.iteration function with Benjamini–Hochberg correction, again using the pSI package for R. For enrichment testing of differentially expressed genes, we restricted analysis to genes that met the following criteria: (1) transcripts for the gene were detected in all eight paired behaving males, and (2) at least 6 transcripts were detected in each subject.
Staging during gastrulation
Cichlid gastrulation was split into three sub stages within the gastrula stage 979. Gastrulation lasts 8 to 12 h, depending on the species, and is defined as after the shield (as described in zebrafish) stage until the presence of the first somite at the beginning of neurula (stage 10). Embryos were classified as early gastrula (EG) by an asymmetry in epiboly after shield stage until the formation of a ridge that is analogous to the anterior neural ridge (ANR) in chick and mouse and the anterior neural border (ANB) in zebrafish. At that point embryos were classified as mid gastrula (MG). MG lasts until the formation of the dorsal–ventral axis, defined by further lengthening of one side of the embryo, which begins to thicken as epiboly progresses. This is the dorsal side of the embryo, and the side opposite the ANR is classified the ventral side of the embryo. At this point the embryos are defined as late gastrula (LG). LG ends with the specification of the neural plate, which appears as a portion of the dorsal embryo that is raised relative to ventral side, usually in line with the ANR.
Immunohistochemical staining
Embryos were harvested at 24 h post fertilization (hpf) from each of the rock-dwelling cichlids Metriaclima patricki, Metriaclima zebra and Labeotropheus fuelleborni as well as the sand-dwellers Aulonocara jacobfreibergi, Copadichromis virginalis, Copadichromis borleyi and Tramitichromis intermedius. The embryos were cultured until they reached gastrula stage, approximately 36 to 40 hpf, then fixed at intervals throughout gastrula until neurula. The embryos were then treated with auto-fluorescence reducer (1.55 mL 5 M NaCl, 250 uL Tris–HCl, pH 7.5, and 95 mg NaBH4) overnight, and 10% 2-mercaptoethanol for 1 h. Next, whole mount in situ hybridization was done, using a modification methods we published previously80. irx1b was visualized using Fast Red (naphthol chromogen, Roche Diagnostics), which fluoresces at near red wavelengths (500–650 nm). After in situ hybridization, embryos were immunostained for pSMAD 1,5,8 protein, using published protocols81. Embryos were then bathed in Vectashield (Vector Labs) containing DAPI and placed in a specially built mold49 that accommodates the large yolk and holds the embryo upright. Embryos were then scanned using a Zeiss LSM 700-405 confocal microscope and processed using LSM 700 software (Zeiss) and Image J82. For experiments in Fig. 2A, multiple embryos of multiple rock- and sand-species were examined (n = 20 embryos total); and in Fig. 2C,D, multiple embryos per gene, genotype and stage were used (n = 110 embryos total).
Rock–sand hybridization and genotyping
Two rock-sand crosses, one between Copadichromis borleyi (CB, sand-dweller sire) and Metriaclima zebra (MZ, rock-dweller dam) and another between Mchenga conophoros (MC, sand-sire) and Petrotilapia sp. ‘thick bar’ (PT, rock-dam), were artificially generated by taking the eggs from the dam just prior to spawning and mixing with sperm from the sire. The resultant F1 were grown in tanks and allowed to spawn normally to generate F2. Several F2 broods were taken from multiple F1 females for each cross, a total of 355 individuals for the CB × MZ cross and 608 for the MC × PT cross. The embryos were fixed at every stage starting at gastrula (stage 9) until early pharyngula (stage 14). The F2 embryos were RNA-extracted at stage 9. DNA extraction was performed by fixing the embryos (stage 11–14) in 70% ethanol, then removing the tail from each individual and extracting the DNA using an extraction kit (Qiagen). Following extraction, the F2 embryos were genotyped using custom probes (CAAATCTCCC[C/T]CCGCGGC, Taqman custom probes, Invitrogen) designed to identify a SNP in irx1b using RT-PCR. A subset of the embryos was also sequenced at a 900 bp interval around the irx1b SNP to verify the custom probes.
Quantitative F2 analysis
We quantified irx1b in F2 at stage 9 and separated by genotypic class. The 74 heterozygous rock X sand F2 embryos were dissected to remove most of the yolk and the total RNA was extracted from each individual using an RNA Extraction Kit (Qiagen).
The amount of mRNA specific to each allele of irx1b was quantified by using the RNA-to-Ct kit (Invitrogen) and the custom probes. The delta Ct for each heterozygote was generated with the equation, 2(allele from dam − allele from sire). We used a Student’s t-test to compare the expression of rock- vs sand-alleles in heterozygous F2.
Forebrain and eye measurements
The telencephalon and forebrain were measured by integrating the area of transverse sections in embryos of rock- and sand-dweller cichlid species, using previously published methods25. The rock-dweller species included Cynotilapia afra (CA, planktivore), Labeotropheus fuelleborni (LF, algivore) and Metriaclima zebra (MZ, generalist); sand-dweller species included Aulonocara jacobfreibergi (AJ, ‘sonar’ hunter), Copadichromis borleyi (CB, planktivore) and Mchenga conophoros (MC, insectivore/generalist). Embryos from each species, as well as the F2 individuals, were measured starting from the earliest the telencephalon can be differentiated from the forebrain (mid-somitogenesis, stage 12) and at each subsequent stage until the forebrain has defined prosomeres (early pharyngula, stage 14)26. To keep measurements standardized across stages, all measurements were defined by forebrain morphology at the earliest timepoint (stage 12). The ‘eye’ measurement remains consistent at all stages, the ‘anterior’ measurement includes the telencephalon and presumptive olfactory bulb, and the ‘posterior’ measurement includes the diencephalon and each of its constitutive prosomeres (dorsal and ventral thalamus and hypothalamus). To facilitate measurements, we used gene expression of rx3 (for stage 12 embryos) and pax6 (stage 13 and 14) to identify the different structures of the forebrain and eye.
RNA extraction and sequencing, adult social behavior
Two adult F1 hybrid males from rock-x sand-interspecific crosses (Supplementary Table S5) were introduced to an assay tank containing 3–5 hybrid females of the same cross with simulated rock habitat on one side and simulated sand habitat on the other side separated by empty tank space (Fig. 3A). Males were observed over a period of four days for courtship behavior, either similar to a rock parent (courting females over the rocky parts of the tank), here designated “social rock”—or to a sand parent (bower building in the sand side of the tank while courting females), here designated “social sand”. When courtship behavior of both males was observed, we primed experiments the next morning by resetting the tanks—knocking down or filling in the sand bower, and adjusting the rock habitat. Both males were allowed to exhibit courtship behavior over a period of 90 min. Males were sacrificed immediately after this 90-min period by rapid decapitation and whole brains were immediately stored in RNAlater (Thermo Fisher Cat# AM7020) within 20 min of decapitation. Two tanks with one F1 male and 3–5 F1 females were also observed. These males exhibited social rock behavior and were designated as “lone” in the behavior assay; their brains were collected in the exact same way as the other samples.
Tissues were frozen in liquid nitrogen, homogenized using a mortar and pestle and placed in trizol. Following standard chloroform extraction, RNeasy mini columns (Qiagen Cat No./ID: 74104) were utilized to purify RNA for sequencing. Total RNA was quantified using Qubit (Molecular Probes) and quality analyzed using the Agilent 2100 Bioanalyzer System for RNA library preparation. RNA input was normalized to 1 µg and libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina-Kit A). Libraries were again quantified, quality assessed, and normalized for sequencing on the HiSeq 2500 Illumina Sequencing System (Georgia Tech Genomics Core, standard practices). Experimental design and raw files can be accessed on the NCBI Gene Expression Omnibus database under the accession number GSE122500.
Differential gene expression analysis
Raw sequence reads from whole brain transcriptomes were quality controlled using the NGS QC Toolkit83. Raw reads with an average PHRED quality score below 20 were filtered out. Filtered reads were also trimmed of low-quality bases at the 3′ end. High quality sequence reads were aligned to the M. zebra reference genome MZ_UMD2a34 using TopHat v2.0.984. On average, across all samples, over 95% of reads mapped to the reference genome. The resulting TopHat2 output bam files were sorted and converted to sam files using samtools v0.1985. Sorted sam files were used as input for the HTSeq-count v0.6.1 program to obtain fragment counts for each locus86. Fragment counts were scale-normalized across all samples using the calcNormFactors function in the edgeR package v3.6.887. Relative consistency among replicates and samples was determined via the Multidimensional scaling (MDS) feature within the edgeR package in R. The native R function hclust(dist) used to cluster samples. Scale-normalized fragment counts were converted into log2 counts per million reads mapped (cpm) with precision weights using voom and fit to a linear model using limma v3.20.988. Pairwise contrasts were constructed between socially rock and socially sand samples. After correcting for multiple comparisons using the Benjamini–Hochberg method89, genes were considered differentially expressed between socially rock and socially sand samples if they exhibited both a fold change ≥ 2 and Padj < 0.05. Using Treefam based mapping69 the cichlid gene names were converted to human analogs and functional enrichment was determined using the TOPPFUN web-browser70.
Ethics statement
All of the animals were handled according to methods approved by the Georgia Tech Institutional Animal Care and Use Committee (IACUC) protocols (permit A100029) with every effort made to minimize suffering and is in compliance with the ARRIVE guidelines.
Supplementary Information
Acknowledgements
This work was supported by Grants from the NIH (R01GM101095, 2R01DE019637-10 to JTS and F32GM128346-01A1 to ZVJ) and the Human Frontiers Science Program (RGP0052/2019 to JTS). We would like to thank Shweta Biliya and the Genomics Core at Georgia Tech for invaluable assistance with the NGS sequencing. We also thank the members of the Streelman lab for comments on this manuscript.
Author contributions
C.P., J.B.S., K.A. and J.T.S. designed research; C.P., J.B.S., K.A., M.W.N., K.P. and R.F.B. performed research; C.P., K.A., M.M., Z.V.J. and P.T.M. analyzed data; C.P., J.B.S., Z.V.J. and J.T.S. wrote the paper. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92385-8.
References
- 1.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nat. Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- 3.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:9718–9723. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 6.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streelman J, Peichel CL, Parichy D. Developmental genetics of adaptation in fishes: The case for novelty. Annu. Rev. Ecol. Evol. Syst. 2007;38:655–681. doi: 10.1146/annurev.ecolsys.38.091206.095537. [DOI] [Google Scholar]
- 8.Colosimo PF, et al. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:E109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kratochwil CF, et al. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science. 2018;362:457–460. doi: 10.1126/science.aao6809. [DOI] [PubMed] [Google Scholar]
- 10.Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro MD, et al. Genomic diversity and evolution of the head crest in the rock pigeon. Science. 2013;339:1063–1067. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamichhaney S, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 13.Okhovat M, Berrio A, Wallace G, Ophir AG, Phelps SM. Sexual fidelity trade-offs promote regulatory variation in the prairie vole brain. Science. 2015;350:1371–1374. doi: 10.1126/science.aac5791. [DOI] [PubMed] [Google Scholar]
- 14.Bendesky A, et al. The genetic basis of parental care evolution in monogamous mice. Nature. 2017;544:434. doi: 10.1038/nature22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfenning AR, et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenwood AK, Wark AR, Yoshida K, Peichel CL. Genetic and neural modularity underlie the evolution of schooling behavior in threespine sticklebacks. Curr. Biol. 2013;23:1884–1888. doi: 10.1016/j.cub.2013.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcovitz A, Jia R, Bejerano G. “Reverse genomics” predicts function of human conserved noncoding elements. Mol. Biol. Evol. 2016;33:1358–1369. doi: 10.1093/molbev/msw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocher TD. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 20.Loh YHE, et al. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh YHE, et al. Origins of shared genetic variation in African cichlids. Mol. Biol. Evol. 2013;30:906–917. doi: 10.1093/molbev/mss326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streelman JT, Albertson RC, Kocher TD. Genome mapping of the orange blotch colour pattern in cichlid fishes. Mol. Ecol. 2003;12:2465–2471. doi: 10.1046/j.1365-294x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- 23.Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parnell NF, Streelman JT. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity. 2013;110:239–246. doi: 10.1038/hdy.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sylvester JB, et al. Competing signals drive telencephalon diversity. Nat. Commun. 2013;4:4. doi: 10.1038/ncomms2753. [DOI] [PubMed] [Google Scholar]
- 26.Sylvester JB, et al. Brain diversity evolves via differences in patterning. Proc. Natl. Acad. Sci. 2010;107:9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baran NM, Streelman JT. Ecotype differences in aggression, neural activity and behaviorally relevant gene expression in cichlid fish. Genes Brain Behav. 2020;19:e12657. doi: 10.1111/gbb.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.York RA, et al. Behavior-dependent cis regulation reveals genes and pathways associated with bower building in cichlid fishes. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E11081–E11090. doi: 10.1073/pnas.1810140115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson ZV, et al. Exploratory behaviour is associated with microhabitat and evolutionary radiation in Lake Malawi cichlids. Anim. Behav. 2020;160:121–134. doi: 10.1016/j.anbehav.2019.11.006. [DOI] [Google Scholar]
- 30.Fraser GJ, et al. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7:e1000031. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streelman JT, Danley PD. The stages of vertebrate evolutionary radiation. Trends Ecol. Evol. 2003;18:126–131. doi: 10.1016/S0169-5347(02)00036-8. [DOI] [Google Scholar]
- 32.Malinsky M, et al. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2018;2:1940–1955. doi: 10.1038/s41559-018-0717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKaye KR, Louda SM, Jay R, Stauffer J. Bower size and male reproductive success in a cichlid fish lek. Am. Nat. 1990;135:597–613. doi: 10.1086/285064. [DOI] [Google Scholar]
- 34.Conte MA, et al. Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. GigaScience. 2019 doi: 10.1093/gigascience/giz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Ari Fuchs S, et al. GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adhikari A, et al. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–185. doi: 10.1038/nature15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesquita LT, et al. New insights on amygdala: Basomedial amygdala regulates the physiological response to social novelty. Neuroscience. 2016;330:181–190. doi: 10.1016/j.neuroscience.2016.05.053. [DOI] [PubMed] [Google Scholar]
- 40.Carvalho FR, Nóbrega CDR, Martins AT. Mapping gene expression in social anxiety reveals the main brain structures involved in this disorder. Behav. Brain Res. 2020;394:112808. doi: 10.1016/j.bbr.2020.112808. [DOI] [PubMed] [Google Scholar]
- 41.Abrahams BS, et al. SFARI gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piñero J, et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Attanasio C, et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rada-Iglesias A, et al. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simoes-Costa M, Bronner ME. Establishing neural crest identity: A gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts RB, Hu Y, Albertson RC, Kocher TD. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13194–13199. doi: 10.1073/pnas.1018456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavodeassi F, Modolell J, Gómez-Skarmeta JL. The Iroquois family of genes: From body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- 48.Bielen H, Houart C. BMP signaling protects telencephalic fate by repressing eye identity and its Cxcr4-dependent morphogenesis. Dev. Cell. 2012;23:812–822. doi: 10.1016/j.devcel.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White DE, et al. Quantitative multivariate analysis of dynamic multicellular morphogenic trajectories. Integr. Biol. 2015;7:825–833. doi: 10.1039/c5ib00072f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volff JN, Korting C, Schartl M. Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol. Biol. Evol. 2000;17:1673–1684. doi: 10.1093/oxfordjournals.molbev.a026266. [DOI] [PubMed] [Google Scholar]
- 51.Ramallo MR, et al. A multidisciplinary study on social status and the relationship between inter-individual variation in hormone levels and agonistic behavior in a Neotropical cichlid fish. Horm. Behav. 2015;69:139–151. doi: 10.1016/j.yhbeh.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Olton DS, Becker JT, Handelmann GE. Hippocampus, space, and memory. Behav. Brain Sci. 1979;2:313–322. doi: 10.1017/S0140525X00062713. [DOI] [Google Scholar]
- 53.Hitti FL, Siegelbaum SA. The hippocampal CA2 region is essential for social memory. Nature. 2014;508:88–92. doi: 10.1038/nature13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang M-C, Huang AJ, Wintzer ME, Ohshima T, McHugh TJ. A role for CA3 in social recognition memory. Behav. Brain Res. 2018;354:22–30. doi: 10.1016/j.bbr.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 55.Zou D, et al. DREADD in parvalbumin interneurons of the dentate gyrus modulates anxiety, social interaction and memory extinction. Curr. Mol. Med. 2016;16:91–102. doi: 10.2174/1566524016666151222150024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connell LA, Hofmann HA. Evolution of a vertebrate social decision-making network. Science. 2012;336:1154–1157. doi: 10.1126/science.1218889. [DOI] [PubMed] [Google Scholar]
- 57.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 58.Menuet A, Alunni A, Joly JS, Jeffery WR, Retaux S. Expanded expression of Sonic Hedgehog in Astyanax cavefish: Multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- 59.Brenner S. Genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernald RD, Maruska KP. Social information changes the brain. Proc. Natl. Acad. Sci. U.S.A. 2012;109(Suppl 2):17194–17199. doi: 10.1073/pnas.1202552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Briscoe SD, Ragsdale CW. Evolution of the chordate telencephalon. Curr. Biol. 2019;29:R647–R662. doi: 10.1016/j.cub.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nazareno AG, Bemmels JB, Dick CW, Lohmann LG. Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Mol. Ecol. Resour. 2017;17:1136–1147. doi: 10.1111/1755-0998.12654. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee TH, Guo H, Wang XY, Kim C, Paterson AH. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strimmer K. fdrtool: Versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 68.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramakrishnan Varadarajan A, Mopuri R, Streelman JT, McGrath PT. Genome-wide protein phylogenies for four African cichlid species. BMC Evol. Biol. 2018;18:1. doi: 10.1186/s12862-017-1072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harris, R. S. Improved Pairwise Alignment of Genomic DNA Doctor of Philosophy thesis, The Pennsylvania State University (2007).
- 72.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 2004;21:468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 75.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grote S, Prufer K, Kelso J, Dannemann M. ABAEnrichment: An R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics. 2016;32:3201–3203. doi: 10.1093/bioinformatics/btw392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murata Y, et al. Allometric growth of the trunk leads to the rostral shift of the pelvic fin in teleost fishes. Dev. Biol. 2010;347:236–245. doi: 10.1016/j.ydbio.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 80.Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. Bmc Biol. 2008 doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tucker JA, Mintzer KA, Mullins MC. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev. Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel RK, Jain M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7:7. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anders S, Pyl PT, Huber W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.