Figure 2. Domain structure of IDA HCs in Chlamydomonas.
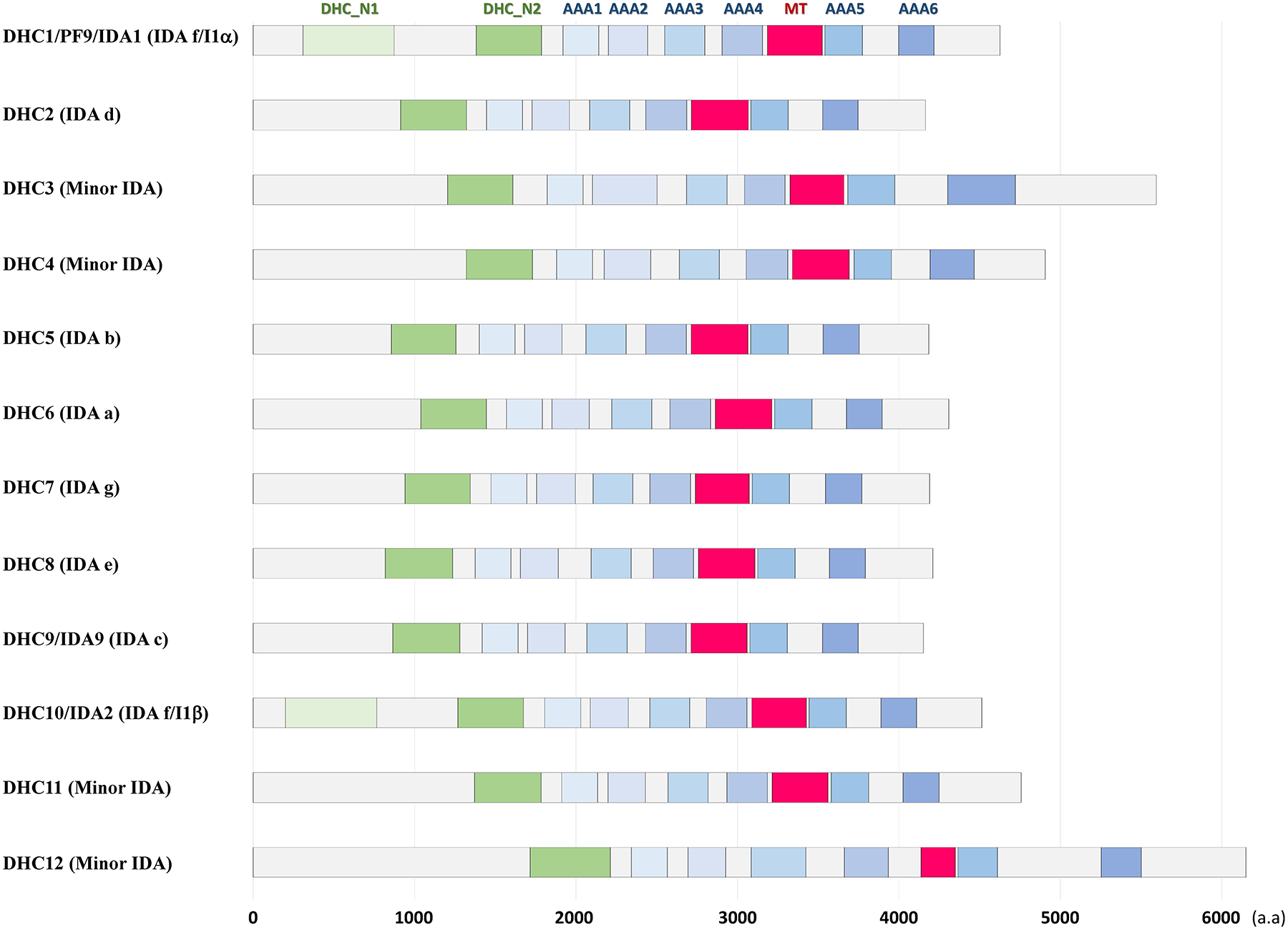
Domains/motifs in DHCs of Chlamydomonas ciliary IDAs. “Dynein heavy chain, N-terminal region 1 (DHC_N1)”, “dynein heavy chain, N-terminal region 2 (DHC_N2)”, and “microtubule-binding stalk of dynein motor (MT)” domains were predicted using the pfam analyses (https://pfam.xfam.org/)(Punta et al., 2012). Appropriate locations of 6 “ATPases-associated-with-diverse-cellular-activities (AAA)” domains/rings in the IDA DHCs were predicted by aligning the IDA sequences with the Tripneustes gratilla ODAβ DHC [X59603.1 (NCBI)](Mocz & Gibbons, 2001). Note that two minor IDAs, DHC3 and DHC12, have longer molecular length than other IDAs. The accession Nos/IDs in the Phytozome Chlamydomonas v5.5 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii) used for these domain predictions were as follows: DHC1/PF9/IDA1, Cre12.g484250.t1.1; DHC2, Cre09.g392282.t1.1; DHC3, Cre06.g265950.t1.1; DHC4, Cre02.g107350.t1.1; DHC5, Cre02.g107050.t1.1; DHC6, Cre05.g244250.t1.2; DHC7, Cre14.g627576.t1.1; DHC8, Cre16.g685450.t1.1; DHC9/IDA9, Cre02.g141606.t1.1; DHC10/IDA2, Cre14.g624950.t1.1; DHC11, Cre12.g555950.t1.2; DHC12, Cre06.g297850.t1.1.
