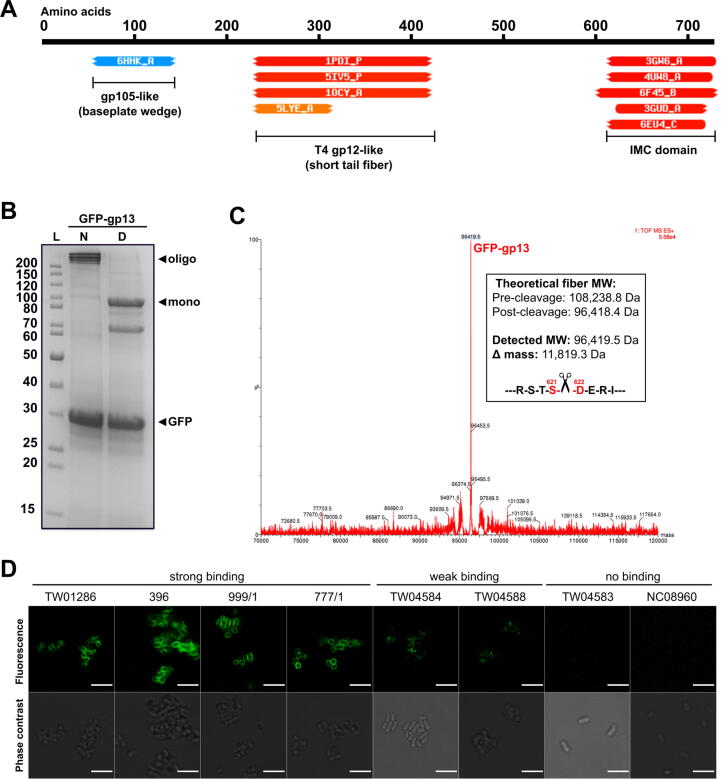
Fig. 1.
Identification and characterization of the tail fiber (gp13) of phage EP335. A) HHpred analysis [47] identified regions of similarity with other phage RBP structures. B) SDS-PAGE of Ni-NTA-purified GFP-gp13. Under native conditions (N; non-boiled), GFP-gp13 formed SDS-resistant oligomers, as observed with other phage RBPs. Heat denaturing (D; 96 °C, 8 mins) caused dissociation into the monomeric form (96.4 kDa; after intermolecular chaperone cleavage). In both samples a GFP contaminant band (~30 kDa) was observed. C) ESI-LC-MS spectra of GFP-gp13 detected a MW of 96,419.5 Da, which corresponds with auto-proteolysis of the C-terminal IMC domain after Ser621. D) Fluorescence and phase contrast images of GFP-gp13 cell binding to different E. coli strains. Cell binding correlated with fluorescence spectroscopy in SupplementaryFig. 2. Scale bars represent 5 µm.