Extended Data Fig. 6.
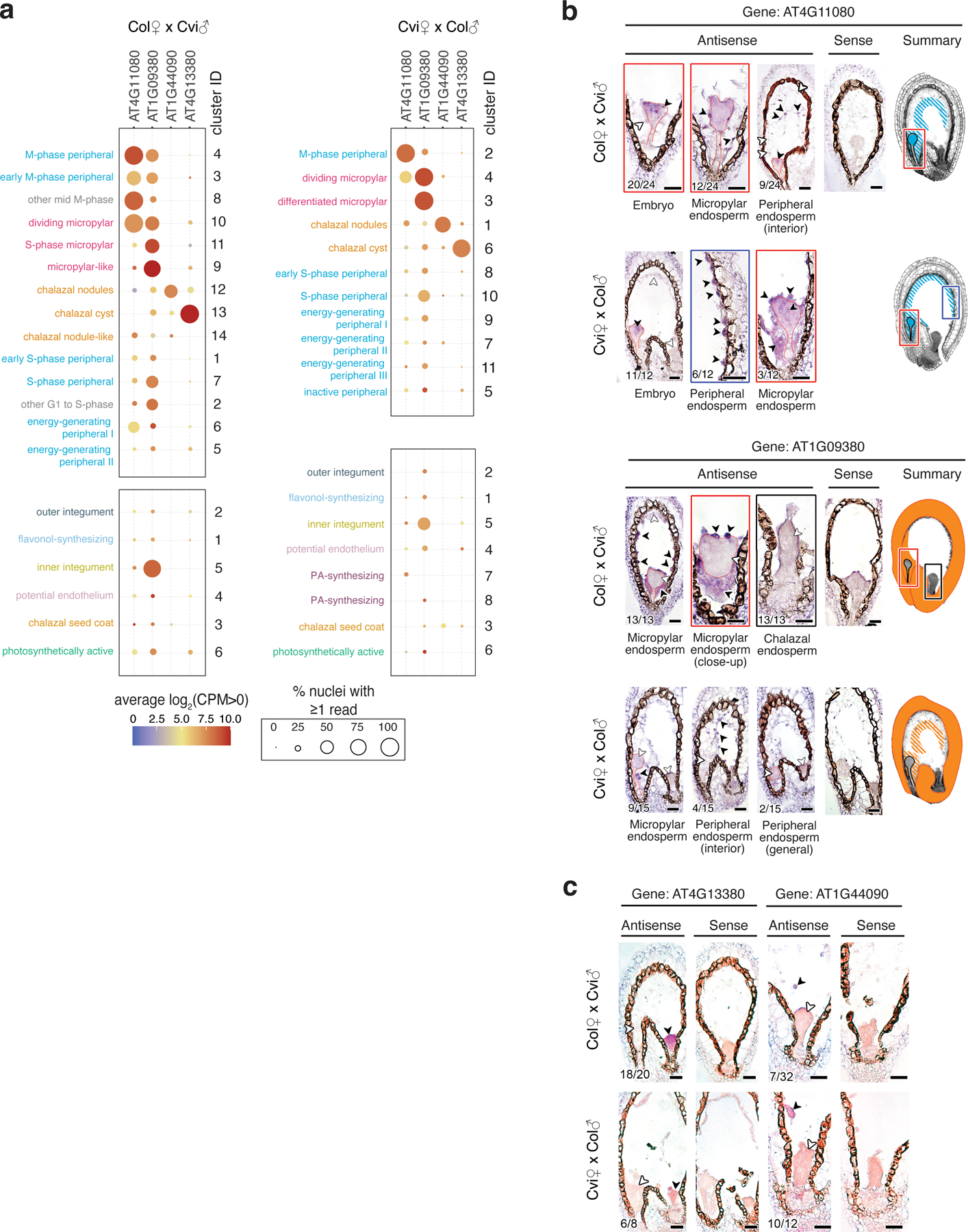
In situ hybridization analysis for additional cluster-specific transcripts.
(a) Expression data for four additional marker genes used for RNA in situ hybridization experiments, across endosperm and seed coat clusters. (b) In situ hybridization (purple signal) results for two micropylar/peripheral clusters. AT4G11080 is most notably expressed in peripheral and micropylar endosperm and in the embryo. AT1G09380 is most notably expressed in the micropylar endosperm and seed coat. In gene summaries, expression indicated by hatched pattern indicates inconsistent expression in that zone among seeds. (c) In situ hybridization results for two additional chalazal endosperm transcripts not shown in Fig. 2: AT4G13380 is predominantly expressed in the chalazal cyst, while AT1G44090 is predominantly expressed in the chalazal nodules. (b-c) Black arrowheads indicate sites of transcript accumulation; white arrowheads indicate examples of sites without transcripts. Number of seeds with expression in specific zones relative to the number of seeds examined is shown in bottom left of panels; expression in one zone does not exclude expression in other zones. Seeds were from three independent controlled pollination events, collected together. For all antisense probes, in situ experiment was performed at least twice, except for AT4G11080, which was performed once. Both sense and antisense probe images shown. Scale bars = 25 μm.
