Fig. 4. Autophagy induction limits SARS-CoV-2 growth in vitro.
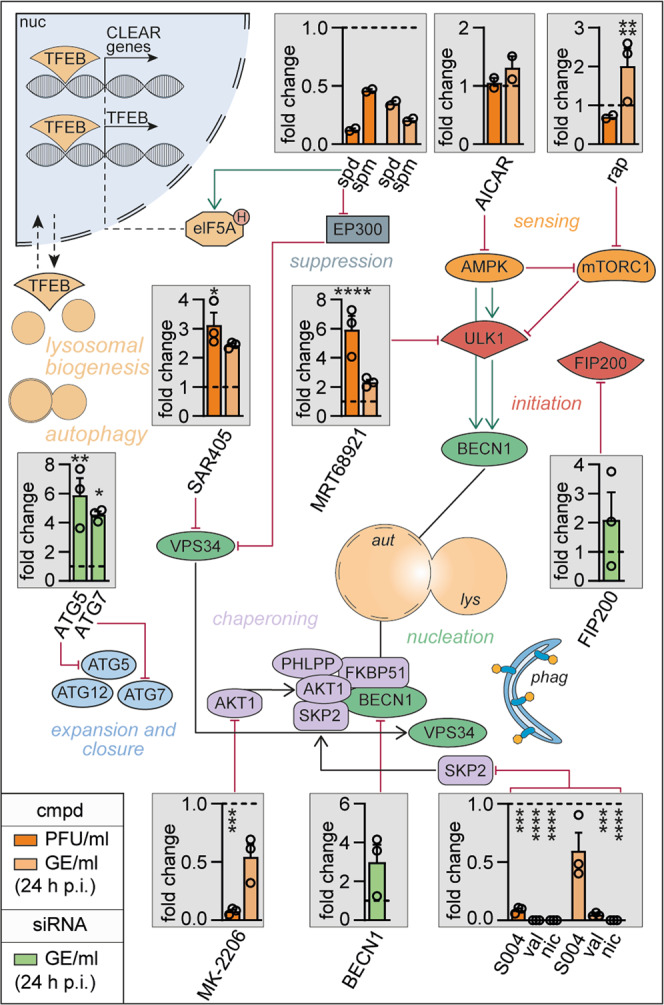
Schematic representation of autophagy signaling indicating site of action of small-molecule compounds (orange columns) or siRNA knockdown (green columns) used for pathway modulation. VeroFM cells were infected with SARS-CoV-2 (MOI = 0.0005) and treated with the indicated compounds: polyamines spermidine (spd, 100 µM) and spermine (spm, 100 µM), AMPK modulator AICAR (25 µM), mTORC1 inhibitor rapamycin (rap, 300 nM), VPS34 and ULK1 inhibitors SAR405 (1 µM) and MRT68921 (5 µM), AKT1 inhibitor MK-2206 (1 µM), BECN1-stabilizers SMIP004 (S004, 10 µM), valinomycin (val, 5 µM), niclosamide (nic, 10 µM) or DMSO (vehicle, dashed lines). For siRNA knockdown, VeroFM cells were transfected with 60 nM siRNA targeting ATG5, ATG7, FIP200, BECN1, and infected with SARS-CoV-2 48 h later. SARS-CoV-2 infectious virus particle units (PFU) and genome equivalents (GE) per ml were determined by plaque assay and real-time RT-PCR at 24 h p.i., respectively. Data are presented as fold differences (see Supplementary Fig. 11 for unprocessed data). In all panels, error bars =SEM derived from n = 3 (n = 2 for spd, spm, AICAR) biologically independent samples from one experiment. Statistics were done for experiments with n = 3 using a two-way ANOVA, Dunnett’s, and one-way ANOVA for siRNA knockdown experiments (green). Compounds with significant SARS-CoV-2 inhibition (spm, spd, MK-2206, S004, val, nic) in VeroFM cells were confirmed in Calu-3 cells (see Supplementary Fig. 11f–g). p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), p ≤ 0.0001 (****), p > 0.05 (not significant, ns). Abbreviations: AICAR, 5-Aminoimidazole-4-carboxamide ribonucleotide; AKT1, RAC-alpha serine/threonine-protein kinase; AMPK, AMP-activated protein kinase; ATG5, autophagy-related 5; ATG7, ubiquitin-like modifier-activating enzyme ATG7; ATG12, ubiquitin-like protein ATG12; aut, autophagosome; BECN1, Beclin-1 protein; CLEAR, coordinated lysosomal expression and regulation; cmpd, compound; EP300, histone acetyltransferase p300; elF5AH, eukaryotic translation initiation factor 5A, hypusinated; FIP200, FAK family kinase-interacting protein of 200 kDa; FKBP51, 51 kDa FK506-binding protein; GE, genome equivalents; lys, lysosome; mTORC1, mechanistic target of rapamycin complex 1; nic, niclosamide; PFU, plaque-forming units; phag, phagophore; PHLPP, PH domain leucine-rich repeat-containing protein phosphatase; rap, rapamycin; S004, SMIP-004; SKP2, S-phase kinase-associated protein 2; spd, spermidine; spm, spermine; TFEB, transcription factor EB; ULK1, unc-51-like kinase 1; val, valinomycin; VPS34, phosphatidylinositol 3-kinase catalytic subunit type 3.
