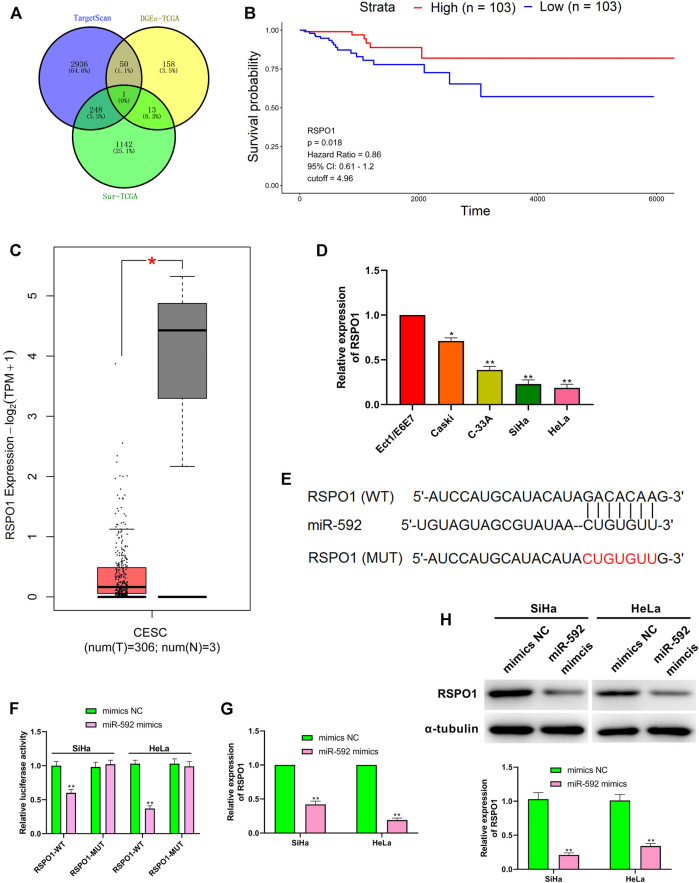
FIGURE 5.
RSPO1 was the target of miR-592. (A) TargetScan, the differentially expressed mRNA screened from TCGA database and the mRNA with significant survival differences screened from TCGA database are used to predict RSPO1 can bind to miR-592; (B) Kaplan-Meier survival curve from TCGA described the overall survival (the criteria of two groups was Quartile); (C) The expression of RSPO1 in CC tissues compared with the normal tissues in the GEPIA website; (D) qRT-PCR was performed to detect the RSPO1 expression in cervical cancer cell lines (Caski, C-33A, SiHa, and HeLa) and normal cervical cell (Ect1/E6E7); (E) TargetScan predicts the binding sites between miR-592 and RSPO1; (F) luciferase activity of RSPO1-wt, RSPO1-mut upon the transfection of miR-592 mimics or mimics NC was evaluated by luciferase reporter assay; (G) The mRNA expression of RSPO1 was detected in SiHa and HeLa cells by qRT-PCR after transfection with miR-592 mimics; (H) The protein expression of RSPO1 was detected in SiHa and HeLa cells by qRT-PCR after transfection with miR-592 mimics. Measurement data were expressed as mean ± standard deviation. The cell experiment is repeated three times independently. *p < 0.05, compared with normal tissues (C); *p < 0.05, **p < 0.01, compared with Ect1/E6E7 group (D); **p < 0.01, compared with mimics NC group (F–H).