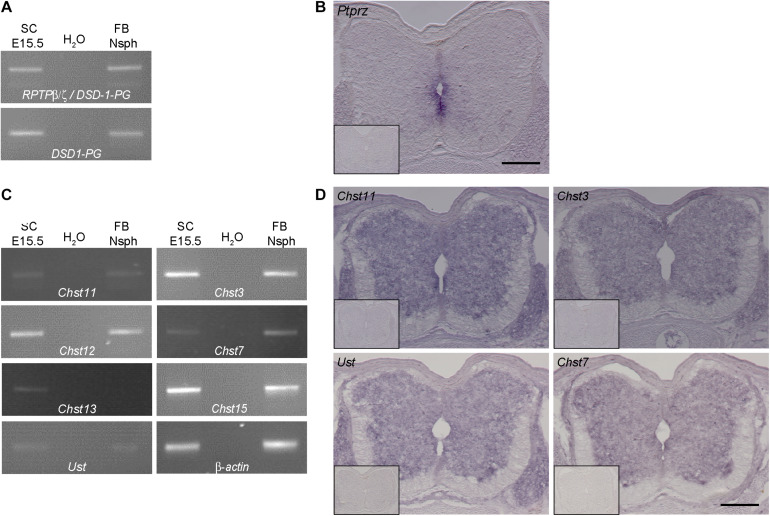
FIGURE 2.
Expression analysis of RPTP-β/ζ /DSD-1-PG and CS-/DS-sulfotransferases in the developing spinal cord. (A) RT-PCR analysis revealed the presence of DSD-1-PG/phosphacan and RPTP-β/ζ mRNAs in tissue isolated from E15.5 spinal cords (E15.5 SC). The isoforms could be detected individually using primers located in the specific sequences of the mRNA (Garwood et al., 1999). (B) The mRNA of the ptprz1 gene coding for a constant part of the RPTP-β/ζ and DSD-1 proteoglycan sequences was produced by cells surrounding the central canal, as could be seen by in situ hybridizations. Sections that were incubated with the sense riboprobe did not show any positive signals (insert). (C) Chst11, Chst12, Chst13, Chst3, and Chst7 as well as Chst15 and Ust genes could be detected in E15.5 spinal cord mRNA via RT-PCR. A representative spinal cord sample is shown together with the no-template control (H2O) and a positive control consisting of cDNA from neurospheres derived from embryonic brain tissue (Akita et al., 2008). Interestingly, although not present in telencephalic mRNA Chst13 could be detected in E15.5 spinal cord tissue. β-Actin served as the reference gene. (D) Representative in situ hybridizations on E15.5 spinal cord sections showed positive signals for Chst3, Chst7, Chst11, and Ust in the central and intermediate zones of the spinal cord, but not in the marginal layer. Sulfotransferase message could be detected in the ventral as well as the dorsal spinal cord and the DRG. Sense controls (inserts) did not show detectable signals. Scale bars: 200 μm.