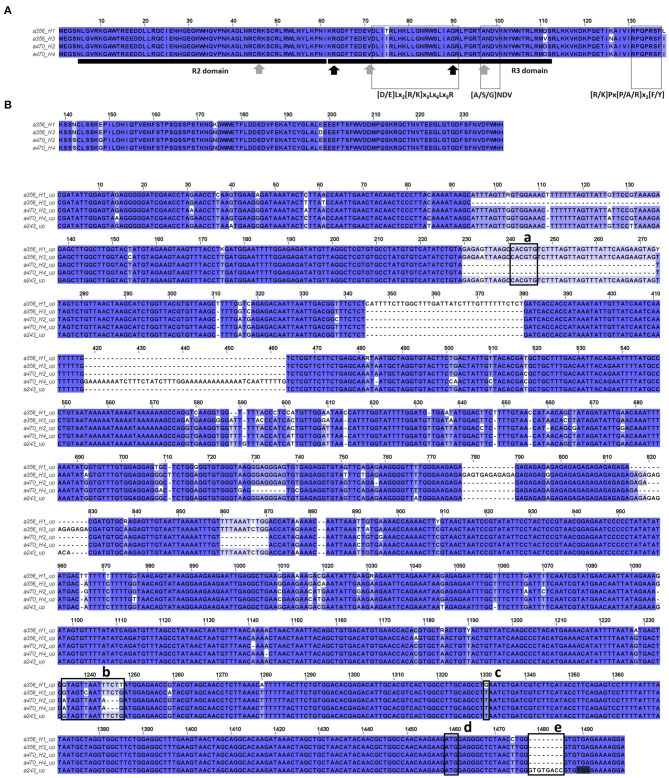
Figure 5.
(A) Alignment of PsMYB10.1 in silico translated proteins from a whole-gene sequence. Conserved amino acids (gray arrows) and signature motifs ([A/S/G]NDV; [R/K]Px[P/A/R]x2[F/Y]) from R2R3-MYB anthocyanin-promoting proteins from Rosaceae were detected in all PsMYB10.1 sequences (Lin-Wang et al., 2010). Conserved motif [D/E]Lx2[R/K]x3Lx6Lx3R indicates a possible interaction with basic-helix-loop-helix (bHLH) proteins. Black arrows indicate key residues in PpMYB10.1 function (Zhou et al., 2018). (B) Polymorphisms found between a356H1, a356H3, a470H2, a470H4, and a243 upstream sequences: (a) G-box motif in the 44 bp indel; (b) polymorphisms in the R1-motif (Espley et al., 2009); (c) SNP S3_12879559 (Salazar et al., 2017); and (e) 8 bp insertion in exon 1 of a243, 4 bases before an in-frame STOP codon (shadowed). The start codon is marked as (d).