FIGURE 1.
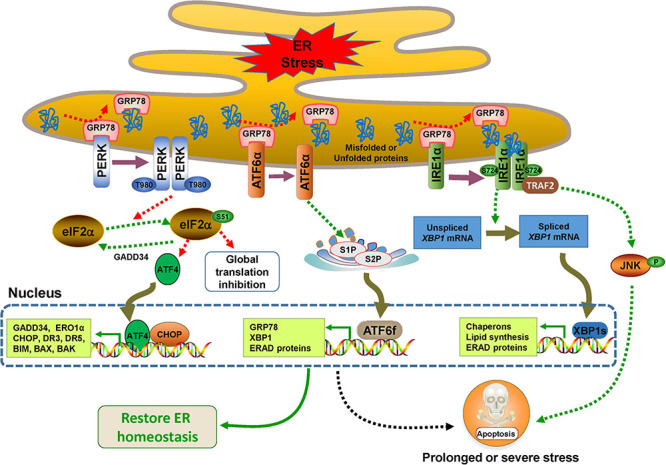
Endoplasmic reticulum (ER) stress and UPR signaling pathways. The UPR is controlled by three major branches, IRE1α, PERK, and ATF6, which bind to GRP78 in the ER under normal conditions. In response to ER stress, the three sensors are activated by dissociating with GPR78. PERK undergoes dimerization, autophosphorylation, and then decreases protein synthesis by phosphorylating eIF2α at the Ser51, which selectively results in the translation of transcription factor 4 (ATF4), a factor that activates transcription of its downstream UPR genes such as CCAAT/enhancer binding protein (C/EBP) homologous protein (CHOP) (Harding et al., 2000). CHOP and ATF4 can upregulate the expression of genes involved in the UPR and apoptosis such as BIM, BAX, BAK, death receptor 3 (DR3), DR5, inositol 1,4,5-trisphosphate (IP3) receptor 1 (IP3R1) and ER oxidase 1α (ERO1α) (Iurlaro and Munoz-Pinedo, 2016). When the ER stress is relieved, CHOP and ATF4 induce the expression of growth arrest and DNA damage inducible protein 34 (GADD34), which directly dephosphorylates eIF2α and restarts global mRNA transcription (Novoa et al., 2001). IRE1α also undergoes dimerization, and even oligomerization and autophosphorylation, and then, its RNase activity is activated and XBP1 mRNA is cleaved to generate XBP1s. The transcription factor XBP1s is responsible for the expression of a subset of downstream genes involved in ERAD, lipid synthesis, protein folding, translocation, and secretion (Korennykh and Walter, 2012; Hetz and Papa, 2018). Activated IRE1α also interacts with tumor necrosis factor receptor (TNFR)-associated factor-2 (TRAF2) and promotes a cascade of phosphorylation events that ultimately activates Jun amino-terminal kinase (JNK)-mediated cell death (Urano et al., 2000). The dissociation of GRP78 drives ATF6 to translocate to the Golgi, where it is cleaved by site-1 protease (S1P) and S2P to generate ATF6f, which transcriptionally activates the expression of a variety of genes involved in ERAD and ER chaperones including GRP78 and XBP1 (Haze et al., 1999; Shen et al., 2002; Yamamoto et al., 2007; Hillary and FitzGerald, 2018).
