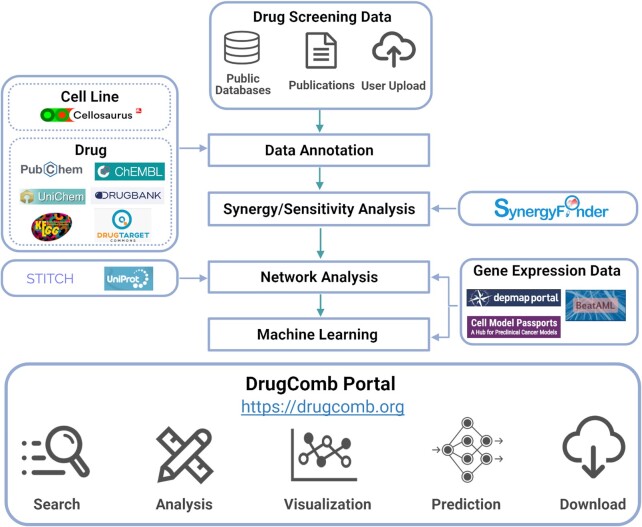
Figure 1.
A schematic overview of the DrugComb database and web server pipeline. Drug combination and monotherapy drug screening datasets are curated from public databases, publications or user-upload. After quality control and pre-processing, the cell information is retrieved from Cellosaurus (23), while the drug information is retrieved from multiple databases including PubChem (24), ChEMBL (25), UniChem (26), DrugBank (27), KEGG (28) and DrugTargetCommons (29). The degree of synergy in drug combinations, as well as the sensitivity of drug combinations and single drugs are determined using the SynergyFinder R package (3). For inferring the mechanisms of action of drugs or drug combinations, their targets as well as interacting proteins are visualized in a signalling network, retrieved from STITCH (30) and UniProt (31). Furthermore, the gene expressions of these proteins in the given cancer cells are obtained from DepMap (32) and Cell Model Passports (33), and from BeatAML where the cancer samples were derived from AML (Acute Myeloid Leukaemia) patients (5). Machine learning algorithms utilize chemical structural and gene expression features to predict drug combination synergy and sensitivity. The DrugComb portal enables the query and download of curated raw datasets and analysis results, as well as the contribution of new datasets.