Abstract
POLE mutations are a major cause of hypermutant cancers, yet questions remain regarding mechanisms of tumorigenesis, genotype-phenotype correlation, and therapeutic considerations. In this study, we establish mouse models harboring cancer-associated POLE mutations P286R and S459F, which cause rapid albeit distinct time to cancer initiation in vivo, independent of their exonuclease activity. Mouse and human correlates enabled novel stratification of POLE mutations into 3 groups based on clinical phenotype and mutagenicity. Cancers driven by these mutations displayed striking resemblance to the human ultra-hypermutation and specific signatures. Furthermore, Pole-driven cancers exhibited a continuous and stochastic mutagenesis mechanism, resulting in inter- and intratumoral heterogeneity. Checkpoint blockade did not prevent Pole lymphomas, but rather likely promoted lymphomagenesis as observed in humans. These observations provide insights into the carcinogenesis of POLE-driven tumors and valuable information for genetic counselling, surveillance, and immunotherapy for patients.
Keywords: Polymerase-exonuclease deficiency, hypermutant cancers, immune checkpoint inhibition, mouse models of cancer, immunotherapy
INTRODUCTION
Eukaryotic DNA replication is a highly accurate process with an error rate of 10−10 mutations per base per cell division[1]. This is enabled by the replication repair machinery which consists of two components. DNA polymerases δ (Pol δ) and ε (Pol ε) are the only nuclear DNA polymerases capable of highly accurate, processive DNA synthesis and current models place Pol δ and Pol ε at the lagging and leading strands, respectively[2–6]. These polymerases replicate the vast majority of the ~3 × 109 basepairs present in the human genome, which must be copied with high fidelity to preserve the transmission of sequence integrity to cell progeny. Two enzymatic properties intrinsic to Pol δ and ε that help to accomplish this task include nucleotide selectivity and 3’ to 5’ exonuclease proofreading.
Should a polymerase incorrectly insert a nucleotide and fail to excise the mispaired base, the mismatch repair (MMR) system can correct these polymerase errors in a post-replicative manner[7]. Complete inactivation of MMR occurs somatically in several tumor types[8] and as germline events in constitutional mismatch repair deficiency (CMMRD)[9] and Lynch Syndrome (LS)[10]. Thus, MMR inactivation is a well-established pathway to mutagenesis and cancer susceptibility. Mutations in POLE are present in multiple cancers with high tumor mutation burdens (TMB)[11–15]. We established the International Replication Repair Deficiency Consortium (IRRDC) to study these replication repair deficient syndromes and tumors in humans. These studies uncovered several key questions regarding POLE tumorigenesis that existing human data cannot sufficiently resolve.
Firstly, only a limited number of POLE mutations cause ultra-hypermutation (>100 mutations per megabase [Mut/Mb]) and cancer, since such mutations must maintain the protein’s polymerase ability while disabling proofreading function[15]. The nature of genotype-phenotype correlation of POLE mutations is therefore poorly understood. Initial reports of cases with germline POLE mutations that disrupt exonuclease activity described a mild phenotype, mainly consisting of adult-onset polyposis[11, 12, 16]. The phenotypic disparity between this benign polyposis syndrome and more aggressive cancers attributable to either germline[17] or somatic[18] POLE mutations suggests variable mutagenic driver capabilities between these mutations. Understanding the nature of this genotype-phenotype relationship is required to better counsel patients and initiate appropriate surveillance protocols.
Secondly, the process of mutation accumulation and tumorigenesis in POLE-driven cancers is poorly described. Human POLE-driven cancers exhibit a unique mutational signature dominated by three distinct base pair substitutions (BPS) defined by the context of their 5’ and 3’ flanking bases: C>A-TCT, C>T-TCG (COSMIC Signature 10)[19] and T>G-TTT (COSMIC Signature 28)[20, 21]. Various models of POLE proofreading deficiency, including experiments in yeast, human cells in culture, and with purified Pol ε protein in vitro, have independently provided evidence that mutant Pol ε is responsible for portions of the above mutational signature but the signature has not been entirely reconstructed in cancers in vivo[22–24]. Moreover, analysis of such signatures in vivo can be used to identify additional secondary mutagenic processes that act during tumor progression.
Finally, immune checkpoint inhibitors (ICI) have shown dramatic responses in hypermutant cancers including melanoma, lung, and MMR deficient tumors[25–28]. However, it is unclear whether POLE mutant tumors will respond to such therapies. Furthermore, preclinical testing for these drugs is hampered by lack of immunocompetent mouse models with hypermutant cancers.
Therefore, animal models which can robustly mimic the human POLE syndrome and cancer development are urgently needed to answer the questions above and to model response of polymerase mutant hypermutant cancers to immunotherapy.
Two previously reported models revealed striking differences in phenotype and failed to answer the above questions. Albertson and colleagues generated a mouse model of Pol ε exonuclease deficiency by engineering double-alanine substitutions at catalytic residues, D272 and E274 (exo−) in the Pole Exo I motif[29]. Strikingly, Pole+/exo- mice do not display any tumor-susceptibility or abnormal survival despite global, germline expression of the exo− allele, which retains negligible exonuclease activity at steady-state[23, 30]. In contrast, recently reported, LSL-PoleP286R mice that conditionally express the most recurrent POLE variants in human cancer, rapidly develop a diverse array of ultra-hypermutated tumors, indicating that the tumorigenic potential of these Pole mutations is variable[31]. These studies did not discuss the mutagenic processes throughout cancer development or their therapeutic implications. Moreover, tumors from LSL-PoleP286R mice did not recapitulate expected Pole-related signatures in their entirety.
To address the tumorigenic and therapeutic potential for POLE cancer variants in vivo, we developed CRISPR/Cas9-mediated knock-in mice harboring germline P286R and S459F mutations. The P286R and S459F alleles have been identified in multiple independent human tumors, with the former representing the most common mutations observed in human POLE driven cancers[32, 33] and the latter is commonly observed as a second somatic occurrence in MMR deficient cancers associated with extremely aggressive cancers with the highest TMBs[18]. Our models exhibit exceptionally strong, yet different, reduction in survival independent of the degree of exonuclease activity providing insight on human and mouse genotype-phenotype. Detailed analysis of these tumors revealed a complete recapitulation of Pole-related mutational signatures and a continuous, stochastic accumulation of mutations resulting in a high degree of inter and intratumoral heterogeneity affecting tumor propagation and treatment.
METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed and will be fulfilled by co-lead contact Uri Tabori (uri.tabori@sickkids.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
PoleS459F Mouse Model
PoleS459F mice were generated in house at The Centre for Phenogenomics (TCP) (Toronto, ON, Canada) by direct delivery of Cas9 reagents to C57BL/6J (The Jackson Laboratory, Stock 000664) mouse zygotes at TCP as performed previously[34]. Briefly, a single guide RNA (sgRNA) with the desired spacer sequence (5’-AGCATCTGACACTGAGTAAG-3’) was synthesized by in vitro transcription from a PCR-derived template. A microinjection mixture of 20 ng/μL Cas9 mRNA (ThermoFisher, A29378), 10 ng/μL sgRNA, and 10 ng/μL single-stranded oligodeoxynucleotide (ssODN) template (5’-ATGAATTTCCTTGGACCCTTGGTTTTTAATGGTCTTGCTCTCTGATGTTCTCCTCAGACTCTGGCtACTTACTtcGTGTCAGATGCTGTGGCTACTTACTACCTGTACATGAAATACGTCCACCCCTTCATATTCGCCCTGTGCA-3’; mutated nucleotides indicated in lowercase) was microinjected into C57BL/6J zygotes. Injected zygotes were incubated in KSOMAA media (Zenith Biotech, ZEKS-50) at 37°C with 6% CO2 until same-day transfer into CD-1 (Charles River Labs, Strain 022) surrogate host mothers. PCR primers (5’- AAACTTGGCTATGACCCTGTAGAG-3’ and 5’-GGGATATCACTTCTGAAGACACCAG-3’) flanking the sgRNA target site and outside of the repair template homology arms, were used to amplify the region of interest from founder progeny. PCR amplicons were subjected to Sanger sequencing. Founders with the desired nucleotide changes were selected for breeding with C57BL/6J mice to produce N1 progeny that were confirmed by sequence analysis of PCR amplicons using the same primers. PoleS459F mice were housed at TCP and all procedures involving animals were performed in compliance with the Animals for Research Act of Ontario and the Guidelines of the Canadian Council on Animal Care. TCP Animal Care Committee reviewed and approved all procedures conducted on animals at TCP.
PoleP286R Mouse Model
PoleP286R mice were generated at The Jackson Laboratory (Bar Harbor, ME, USA) using a CRISPR/Cas9-mediated knock-in strategy. Three overlapping sgRNAs with the desired spacer sequences (5’-TCAGCATCAGGGAATTTGAG-3’, 5’-TCTGATCGGTCTCAGCATCA-3’, 5’-ATCTGATCGGTCTCAGCATC-3’) were synthesized by in vitro transcription. A microinjection mixture of wildtype Cas9, sgRNA, and ssODN (5’-GTCCTTTTAGGACCCTGTGGTTTTGGCATTTGACATCGAGACGACCAAACTGCCTCTCAAATTCCgTGATGCcGAGACCGATCAGATCATGATGATCTCCTATATGATTGATGGCCAGGTGAACAGAATC-3’; mutated nucleotides indicated in lowercase) was microinjected into C57BL/6J zygotes. Injected zygotes were then transferred to pseudopregnant female mice. PCR primers (5’-TCCAAGATGAAGATGTTGTCCC-3’ and 5’-CTAATCCACCCACAAGCCTC-3’) residing outside the of the repair template homology arms and capturing the gRNA target site were used to amplify the region of interest from founder progeny. Sanger-sequencing of PCR amplicons were performed to identify PoleP286R/+ mice, which were selected for breeding with C57BL/6J mice to produced N1 progeny. Genotyping of N1 mice were performed as above. All animal care and procedures were approved by the Tulane School of Medicine Institutional Animal Care and Use Committee (Protocol #4445). All PoleP286R/+ and wildtype control mice were housed at the Tulane University SOM Vivarium (New Orleans, LA, USA).
Mlh1−/− Mouse Model
Mlh1−/− mice[35] were provided by A. Martin (University of Toronto) and housed as indicated above for PoleS459F mice. Mlh1 genotyping was performed as indicated previously[35] using the following primers: primer A, TGTCAATAGGCTGCCCTAGG; primer B, TGGAAGGATTGGAGCTACGG; and primer C, TTTTCAGTGCAGCCTATGCTC.
METHOD DETAILS
Determination of Pol ε Expression
Expression of the PoleS459F mutant allele was assessed in ear notches derived from adult PoleS459F/+, PoleS459F/S459F, and Pole+/+ mice. Total RNA was purified using the RNeasy Mini Kit (Qiagen). cDNA was synthesized using SuperScript® IV Reverse Transcriptase (Thermo Fisher Scientific) on 1μg of RNA. PCR was subsequently performed on cDNA with intron-spanning primers (forward: 5’-ACTGCCTCAGGTGGGTGAAG-3’ and reverse: 5’-CTTCCGCAGCACCTCACTAGG-3’). Products were visualized by agarose gel electrophoresis using GelRed (Biotium). Gel purification was performed followed by Sanger sequencing to detect mutant and WT alleles.
Pathology
Complete necropsies were performed, and the following tissues were taken for histology: brain, liver, spleen, heart, lungs, thymus, small and large intestines, and any abnormal tissue. Tissues samples were fixed in 10% phosphate buffered formalin, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and examined by light microscopy. Pathologists (I. Siddiqui and C. Hawkins) reviewed all slides. Tumors were confirmed histologically.
Immunohistochemistry
Immunostaining was performed on unstained 5 μm slides from FFPE blocks. Sections were deparaffinized, rehydrated, and subjected to heat-mediated epitope retrieval using 0.01M Citrate Buffer pH 6.0. Sections were washed 2× 5 min in PBS-T and, blocked in 2.5% Normal Goat Serum and incubated with either anti-CD3 (Dako, A0452; 1:200 dilution) or anti-B220 (Pharmingen, 553084; 1:2000) primary antibodies for 1 hour at room temperature. Endogenous peroxidases were blocked using BLOXALL Blocking Solution (SP-6000) at room temperature for 15 minutes. Sections were subsequently washed and treated with ImmPRESS (Perixodase) Polymer IgG reagent (either ImmPRESS HRP Anti-Rabbit IgG (Peroxidase) Polymer Detection Kit, Vector Laboratories MP7451 or ImmPRESS HRP Anti-Rat IgG (Peroxidase) Polymer Detection Kit, Vector Laboratories MP-7404) for 30 minutes at room temperature. Sections were washed and then treated with DAB (Vector Laboratories SK-4100) for visualization followed by double distilled water for to stop the reaction. Sections were counterstained with Hematoxylin, dehydrated, mounted with a coverslip using VectaMount Permanent Mounting Media (Vector Laboratories, H-5000).
Literature Search for case reports of germline POLE mutations
MeSH search terms: (((((((“hereditary colorectal cancer”) OR “colorectal cancer”) OR “endometrial cancer”) OR polymerase proofreading associated polyposis) OR “genetic risk factors”) AND POLE) AND mutations) AND germline).
POLE exonuclease excision rate assay
Excision rate constants were measured as previously described[36, 37]. Briefly, a pre-incubated solution of Pol e (100 nM) and 5′−32P-labeled DNA substrate (20 nM) was rapidly mixed with Mg2+ (8 mM) in reaction buffer at 37°C. After various incubation times, the reaction was quenched with the addition of EDTA. The excision rate constants for POLE wild-type, D275A/E277A, P286H, P286R, F367S, L424V, L424I, and S459F were measured using a rapid chemical quench-flow apparatus. Product concentration was plotted versus time and fit to a single-exponential equation, [product] = Aexp(−kexot), to yield the excision rate constant, kexo.
Prophylactic Immunotherapy
PoleS459F/S459F mice were stratified into four treatment groups and beginning at 6–8 weeks of age were injected intraperitoneally (IP) with either: 1. Vehicle-only (PBS, pH = 7.0); 2. Anti-mouse CTLA-4 200μg (BioXcell, Catalog No. 648317J2B); 3. Anti-mouse PD1 250μg (BioXcell, Catalog No. 665418F1); or 4. Anti-mouse CTLA-4 200μg + anti-mouse PD1 250μg. Mice were injected every 3–4 days until endpoint. At endpoint, mice were euthanized and whole-body necropsied. All macroscopically visible abnormal tissue was collected and subsequently divided to formalin-fix and paraffin-embed, snap freeze, and dissociate for viable freezing of tumor cells.
Statistical Analysis
Kaplan-Meier curves of mouse survival logs were generated via the GraphPad Prismv7 software. Log-rank test was performed with the same software to calculate the p-value. Statistical significance was set at p < 0.05.
Whole Exome Sequencing and Variant Calling
DNA from both tumor and normal tissues was isolated using the Qiagen DNeasy Kit (Catalog No. 69504) and subsequently submitted to The Center for Applied Genomics (Hospital for Sick Children, Toronto, ON, Canada), for whole exome sequencing (WES), alignment to the reference genome and variant calling. Agilent’s SureSelectXT Mouse All Exon kit was used for enrichment and paired-end sequencing was done on Illumina HiSeq4000. For two PoleP286R/+ tumors (1098 and 1144), tumor and control tail gDNA was submitted to the Beijing Genomics Institute (BGI) Americas at Children’s Hospital of Philadelphia (CHOP). Prior to sequencing, DNA was enriched using Agilent SureSelect XT Mouse All Exon Kit. Paired-end WES was performed on an Illumina HiSeq4000. FASTQ files from these two mice were processed in the same manner as all other samples in the current study as follows. The software bcl2fastq2 v2.17 was used to generate raw FASTQ files. Alignment to the mouse mm10 reference genome was done using BWA-MEM 0.7.12, followed by PicardTools 1.133 to mark duplicates and sort the BAM file, and GATK 3.4–46 ‘IndelRealigner’ for local realignment of reads. Variant calling was done as described by Doran et al., 2016 using SAMtools 1.3.1 (for mpileup), BCFtools 1.3.1 and VCFtools 0.1.12a[38]. GATK MuTect2 was also used to compare tumor and matched normal samples to call somatic SNVs and indels and annotation of variants was done using ANNOVAR (version Feb 2016).
Data Analysis
The TMB from WES data was plotted per chromosome using easyGgplot2 on R. DeConstructSigs[39] was used to determine COSMIC signatures (Alexandrov et al., 2013) in the mutation spectrum within a tri-nucleotide context for each sample. All analyses were done on R version 3.4.4 using the high-performance computing cluster at the Hospital for Sick Children.
Data Availability
Data generated during our study have been deposited in the NCBI’s Sequence Read Archive (SRA) database under the accession code PRJNA659565.
Mass Cytometry Methods
Cell Staining for Mass Cytometry
Spleen, lymph node or thymus from PoleS459F/+ or PoleS459F/S459F mice showing clinical signs of malignancy and enlargement of one or more lymphoid tissues were mechanically digested to release single cells and cryopreserved. Freshly harvested spleen from Pole+/+ control mice was dissociated into single cell suspensions on the day of each immunophenotyping experiment to use as a staining control. Cryopreserved cells were thawed and washed twice in pre-warmed complete (c) RPMI (RPMI, 10% FBS, 25mM Hepes, 55 μM β-mercaptoethanol, 0.1 mM non-essential amino acids, 1mM sodium pyruvate, 2mM L-Glutamine) containing 200 μg/ml DNAse. Cells were pelleted, resuspended in cRPMI, and counted using Trypan Blue via a hemocytometer. Cell concentration was adjusted to 2×106 cells/ml and placed at 37°C in a humidified 5% CO2 incubator for 30ʹ. After 30’, 500nM 127IdU (5-127Iodo-2’ deoxyuridine) was added for an additional 60ʹ to label newly synthesized DNA in proliferating cells. Cells were then washed prior to blocking Fc Receptors as previously described[40], washed again and then stained for 30’ at RT with anti-CD45 tagged with 89Y or 156Gd. After washing, pairs of bar-coded samples (stained with different CD45 conjugates) were combined into a single 5 mL tube polypropylene to perform multi-plexed staining with pre-determined optimal concentrations of metal-tagged antibodies specific for cell surface markers, Cisplatin (BioVision Inc., USA) to stain dead cells and transcription factor antibodies as previously described[40]. After the final wash, cells were re-suspended in PBS containing 0.3% saponin, 1.6% formaldehyde, and 100 μM 191/193Iridium to stain nuclear DNA for up to 48h at 4°C. Prior to analyzing stained cells on the Helios, cells were washed and re-suspended in Maxpar Cell Acquisition Solution (Fluidigm, Markham ON Canada) at 2–5×105/ml followed by addition of 5-element EQ normalization beads (Fluidigm, Markham ON, Canada). Samples were acquired on the Helios according to Fluidigm’s protocols. The Helios software (v6.7.1014) was used for pre-processing to generate and normalize FCS 3.0 datafiles.
Metal-tagged Antibodies
The following antibodies were directly purchased from Fluidigm: CD45-89Y, CD150-167Er, and Lag3-174Yb. All other antibodies were purified from hybridoma supernatants in-house or purchased as purified carrier-free antibodies and metal-tagged using Fluidigm Maxpar Metal Conjugation Kits according to the manufacturer’s instructions. Panel and antibody information can be found in Table S1. This panel was used in two experiments to stain single cell suspensions from enlarged lymphoid tissues from a total of 16 untreated mice, 4 anti-CTLA-4 treated mice, 5 anti-PD1 treated mice, and 6 combination therapy treated mice.
CyTOF Data Analysis
FCS 3.0 files were uploaded into Cytobank (Santa Clara, CA) and each parameter was scaled using the Arcsinh transformation (scale argument of 5). Each 2-plex FCS file was manually de-convolved into separate FCS files (using the ‘split files by population’ feature in Cytobank) containing either CD45-89Y+ or CD45-156Gd+ cells, which were then further gated to remove EQ beads, dead cells, debris, and aggregates. FCS files containing 12,500 CD45+ live single cells from each sample were exported for clustering using the FlowSOM algorithm (github.com/SofieVG/FlowSOM) [41]. Clustering was performed (k = 30, Arcsinh scale argument of 5) on 2 WT spleen and 16 tumor samples using the markers indicated in Table S1. The FlowFrame utilility within the Bioconductor R package ‘flowCore’ was used to create new FCS files that include the FlowSOM cluster IDs, which were then uploaded to Cytobank where t-SNE dimensionality reduction was performed (iterations: 3000, perplexity:30, theta: 0.5) using the clustering markers. The FlowSOM cluster IDs were also included to enhance visualization of the FlowSOM clusters in the t-SNE embedding[42].
Statistical Analysis for Mass Cytometry Data
Prismv8.2.0 was used to perform ordinary one-way ANOVA with post-hoc comparisons of means of each column with every other column (Fig. 3C–E) or the mean of untreated control with the other columns (Fig. 4C). Correction for multiple testing was performed using the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli (FDR=5%) to calculate adjusted p values (ie., q values). In Fig. 3, 4 and Supplementary Fig. 6, *: q<0.033; **: q<0.002; ***: q<0.001.
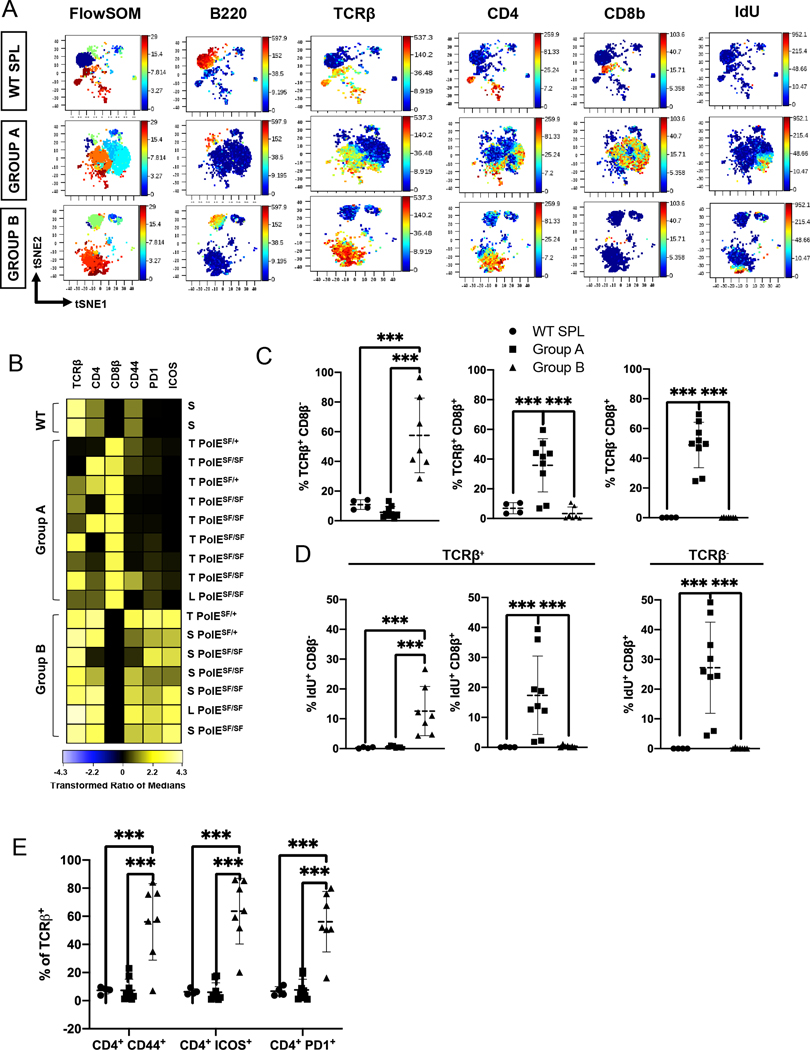
Figure 3. Identification of two distinct types of T cell lymphoma in Pole mutant mice.
(A) Representative t-SNE plots of FlowSOM clusters present in wild-type (WT) SPL (top) compared to representative Group A (middle; 3031) and Group B (bottom; 3158) lymphomas. Maps were colored in the Z dimension by FlowSOM cluster number (left) or the indicated lineage markers.
(B) Heatmap of marker expression (columns) by CD11b− B220− cells in each sample (rows). Marker intensity was normalized to the transformed ratio of medians by the column’s minimum. Samples were manually grouped as control WT SPL (n=2), Group A lymphomas with high CD8β and variable TCRβ and CD4 expression (n=9), and Group B lymphomas with high TCRβ and CD4 and little CD8β expression (n=7) subsets. Sample ID to the right indicates Pole genotype (S459F/S459F = SF/SF; S459F/+ = SF/+) and tissue: thymus (T), spleen (S) and lymph node (L).
(C) Scatter plots show the % of each subset (y axis), identified by manual gating, of the indicated subsets among live single cells: TCRβ+ CD8β− (left), TCRβ+ CD8β+ (middle) and TCRβ− CD8β+ (right) in WT SPL (n=5, circles) versus Group A (n=9, squares) and Group B (n=7, triangles) lymphomas. Means are identified with a dashed horizontal line on each bar and whiskers show the SD. These graphs include all the lymphoma samples shown on the heatmap in part B.
(D) Scatter plots show the percentage of IdU+ CD8β+ and IdU+ CD8β− cells within the TCRβ+ subset (left and middle) compared to the % IdU+ CD8β+ cells in the TCRβ− B220− CD11b− subset (right) defined as shown in Supplementary Fig. 6A. Data are shown for the same samples shown in part C.
(E) Scatter plots configured as described in (C) show the relative abundance of CD4+ cells expressing CD44, PD1 or ICOS cells among TCRβ+ cells from the groups shown in part C. Differences between groups were evaluated by ordinary one-way ANOVA, using a false discovery framework (FDR) of 5% to yield q values. Significant differences are noted by: ***, q<0.001; **, q<0.002, *, q<0.033.
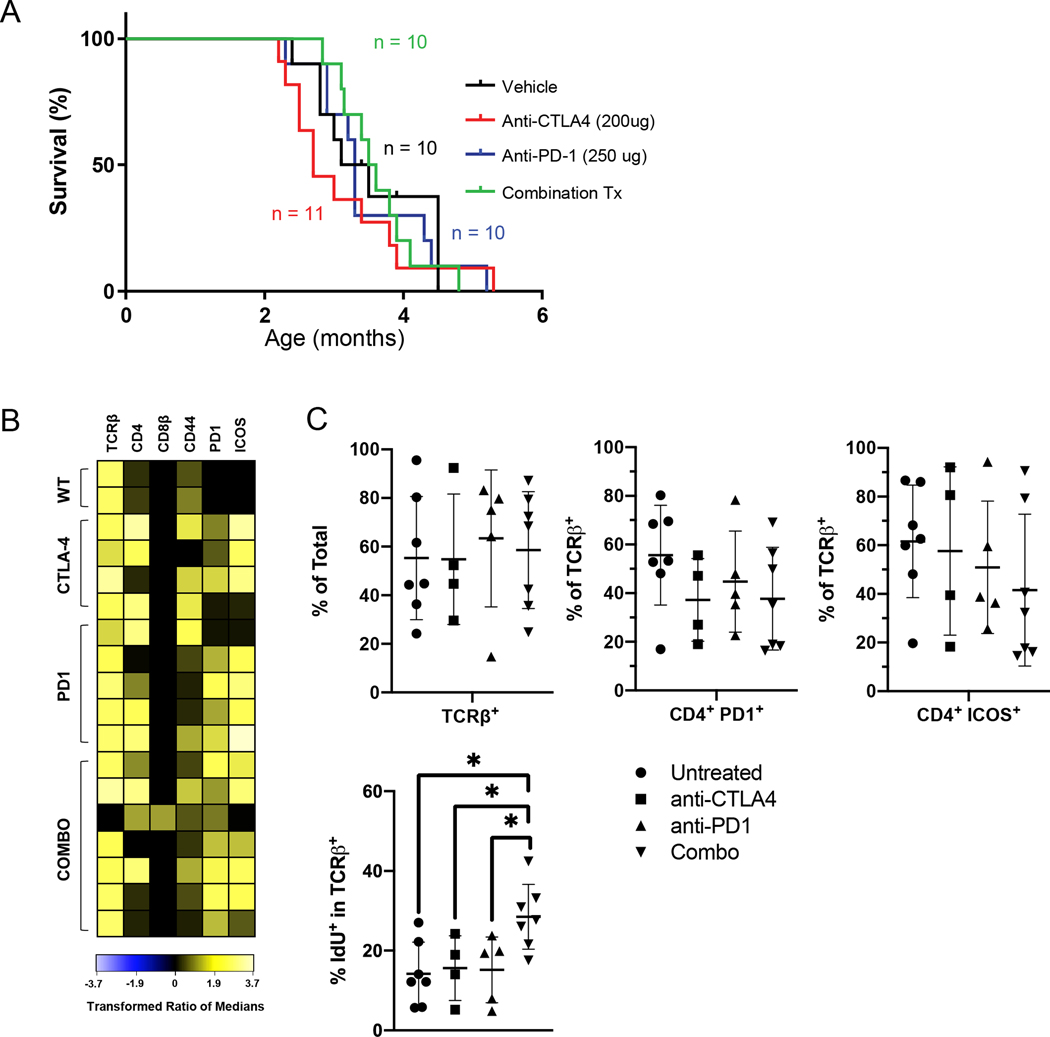
Figure 4. Impact of prophylactic immune checkpoint blockade on Pole driven lymphomagenesis.
(A) Kaplan-Meier survival estimates from control, anti-PD1, anti-CTLA-4, and combination treated PoleS459F/S459F mice. Treatments were delivered intraperitoneally (IP) twice weekly beginning at 6 weeks of age until mice were endpoint. Tumor-free, and overall survival findings are identical. One month = 4.3 weeks. (p = 0.82; Log-rank test).
(B) Heatmap of marker expression (columns) by CD11b− B220− cells in each sample (rows) from from WT SPL or each treatment group at end-point configured as described for Figure 3B. Data for the untreated mice (n=7) was replotted from Figure 3.
(C) Top: Scatter plots (configured as described in Fig. 3) show the percentage of TCRβ+ cells among total live cells (left) and the %CD4+ PD1+ (middle) or %CD4+ ICOS+ cells among TCRβ+ cells in each treatment groups. There was a single Group A tumor in the Combo treatment group in which most proliferating cells were TCRβ− CDβ+ PD1− (not shown), but we chose not to exclude this sample to keep the analysis unbiased. Bottom: Scatter plots show relative the %IdU+ cells in the TCRβ+ subset from each treatment group. Anti-CTLA-4 (n=4), anti-PD1 (n-5), Combo (n=7). Significant differences in abundances were tested and displayed as in Fig. 3 except that each treatment group was compared only to the untreated group.
RESULTS
Pole mutant mice provide insight into genotype-phenotype in humans.
To model the role of different POLE exonuclease domain mutants (EDM) on tumorigenesis in vivo, we used a CRISPR/Cas9-mediated knock-in approach to generate mice harboring the clinically relevant, P286R or S459F substitutions (Supplementary Fig. S1A and S2A). Expression of mutant and WT alleles was confirmed by either sequencing of cDNA amplified using intron-spanning primers (Supplementary Fig. S1B and S2B). Litters from PoleS459F/+ PoleS459F/+ breeders exhibited normal Mendelian distribution of WT and mutant Pole alleles (p = 0.9869, Chi-Square independence test). However, litters from PoleP286R/+ PoleP286R/+ breeders never produced PoleP286R/P286R neonates (p = 0.0014, Chi-Square independence test) and PoleP286R/P286R embryos were not observed after embryonic day E13.5, suggesting embryonic lethality (Supplementary Fig. S1C). In contrast, male and female PoleS459F/+, PoleS459F/S459F, and PoleP286R/+ mice reached sexual maturity and were fertile.
Although all PoleS459F/+, PoleS459F/S459F, and PoleP286R/+ mice survived into adulthood, they rapidly succumbed to aggressive cancers beginning as early as 1.2 (PoleS459F/S459F), 4.1 (PoleS459F/+) and 2.0 (PoleP286R/+) months. Tumor-free survival differed significantly between genotypes (Fig. 1A). As expected, given the additional mutant allele, PoleS459F/S459F mice had significantly shorter median survival compared to PoleS459F/+ mice (p < 0.0001). Moreover, PoleP286R/P286R mice were non-viable, confirming a more severe phenotype in homozygous mutants compared to heterozygous P286R mutants in vivo consistent with previously characterized Pole mutant mice[29, 31]. Importantly, PoleP286R/+ mice had shorter tumor-free survival than PoleS459F/+ mice, and since PoleS459F/S459F were viable, the same could be presumed for homozygous mutants. Furthermore, the influence of a single P286R or S459F allele on survival observed here was remarkably more severe than the Pole exo− allele previously reported[29]. Together, these findings reveal a genotype-phenotype correlation between Pole mutant genotypes and tumor penetrance in mice.
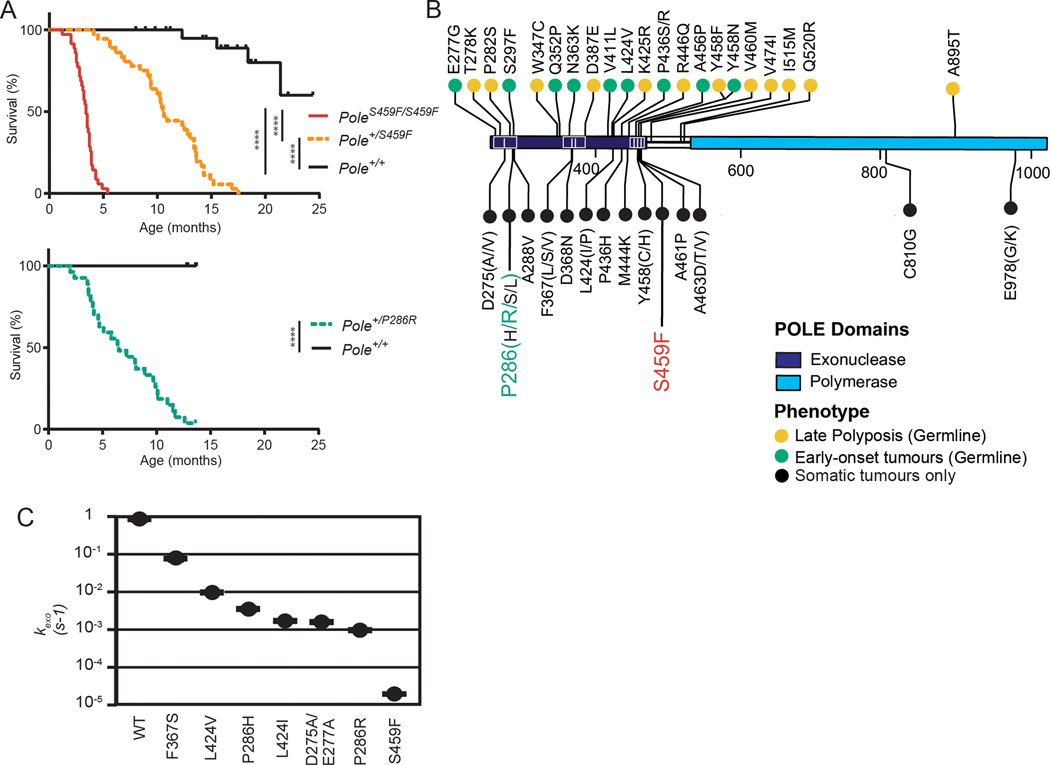
Figure 1. Pole mutations confer variable tumorigenic capabilities in vivo and support a genotype-phenotype correlation.
(A) Kaplan-Meier survival estimates. Mice were followed for long-term survival and observed daily until endpoint. Upper: PoleS459F/S459F (n= 31); PoleS459F/+ (n=33); Pole+/+ (n=16); Lower: PoleP286R/+ (n = 27); Pole+/+ (n=17). One month = 4.3 weeks. Significance are indicated using Log-rank test.
(B) Landscape of germline and somatic POLE driver mutations. Mutations designated as germline were collected from the IRRDC or were previously reported. Mutations designated as “somatic only” are not found in germline cases but were validated drivers as determined by our previous comprehensive characterization[15]. Residues upstream of the exonuclease domains (1–268) and downstream of the polymerase domain (1100 – 2286) are omitted for clarity.
(C) Excision rate constants measured for 7 POLE exonuclease mutants. POLE mutants are indicated on the x-axis. Error fitting for each curve was performed as described previously[37].
Necropsy and pathology findings revealed that the majority of PoleS459F/+, PoleS459F/S459F, and PoleP286R/+ mice had hepatosplenomegaly, large mediastinal (thymic) masses, and/or enlarged lymph nodes, suggesting the presence of lymphomas with similar characteristics to those described in replication repair deficient (RRD) models[43–45] (Supplementary Table S2 and Supplementary Fig. S1D–H and S2C–I). Other tumor types observed in PoleS459F mice included those observed in humans such as adenoma and adenocarcinoma of the gastrointestinal (GI) tract, and other uncommon cancers such as sarcoma, and germ cell.
To determine the relevance of this correlation to human disease, particularly with respect to instances of germline POLE mutations, we collected clinical data from the IRRDC[15, 18] and published data on cases harboring germline or sporadic POLE mutations (summarized in Supplementary Table S3). We found that driver status POLE mutations could be classified into three distinct categories corresponding to their clinical presentation (Fig. 1B). Less severe mutations, including several outside the exonuclease domain, were detected in the germline of individuals across families, had low penetrance, predisposing to a mild phenotype of adult-onset polyposis of the GI tract and late-onset colorectal cancer (CRC). A second set of mutations, including P436R, were also found in the germline and predisposed to early-onset, more aggressive tumors such as brain malignancies and childhood-onset CRC. Finally, a third group of mutations, including P286R and S459F, were only observed as somatically acquired mutations in various cancers suggesting that these mutations are more aggressive than mutations tolerated in the germline.
To explore the potential mechanisms of this phenomenon and its correlation to the human syndrome we first determined whether survival differences between Pole mutant mice could be explained by differing effects of mutations on exonuclease function. We performed in vitro enzymatic reactions as previously conducted to assess exonuclease activity of several POLE mutants[36, 37]. Interestingly, the S459F mutation ablated exonuclease activity nearly 100-fold more than either the P286R or D275A/E277A mutations, suggesting that exonuclease activity alone cannot explain the ability of a mutation to drive tumor development in vivo (Fig. 1C). Recent data in Saccharomyces cerevisiae demonstrate that the mutation rate of P286R is the highest observed, and both P286R and S459F are significantly higher than D275A/E277A[22]. Moreover, the ultramutator phenotype conferred by the P286R mutation may be due to hyperactive polymerase activity rather than decreased exonuclease activity alone[46]. These findings provide possible mechanisms that explain the differences in survival observed between Pole mutants and support a genotype-phenotype correlation of POLE mutations in humans.
The genetic profiles of Pole mutant tumors resemble human cancers
POLE exonuclease deficiency in human malignancy is genetically characterized by genome-wide ultra-hypermutation and mutational signatures 10[19] and 28[20, 21]. However, the kinetics of mutation accumulation over time is not known. To study these processes, we performed whole exome sequencing (WES) on multiple tumors and spatial locations from PoleP286R/+, PoleS459F/+, and PoleS459F/S459F mice. Sequenced tumors mainly consisted of lymphomas (26/29 across all three Pole genotypes) since these were the predominant tumors observed, but also included several non-lymphoma tumors (1 GI, 1 testicular cancer, and 1 sarcoma) (Fig 2A). All Pole mutant tumors were ultra-hypermutant with an average TMB of 170.5 Mut/Mb across all three genotypes (Fig. 2A), comparable to findings from human POLE mutant cancers. To ensure these findings were unique to Pole mutant mouse tumors, we sequenced tumors (5 lymphomas and 1 GI cancer) from MMRD mice harboring homozygous deletion of Mlh1 (Mlh1−/−)[35], as well as a cohort of tumors (5 lymphomas, 2 brain tumors, and 1 lung cancer) from replication repair proficient mice. Mlh1−/− tumors were hypermutant and exhibited mutational burdens consistent with human LS and CMMRD cancers (between 10 to 100 Mut/MB) but were significantly lower than all Pole mutant tumors (p=0.00022). Both Pole mutant and Mlh1−/− mouse tumors harboured significantly higher mutational loads than replication repair proficient mouse cancers (p=2×10−5 and 0.0037, respectively; Fig. 2A). These mutations were evenly distributed throughout the exome (Fig. 2B and Supplementary Fig. S3) suggesting a stochastic mutagenesis mechanism.
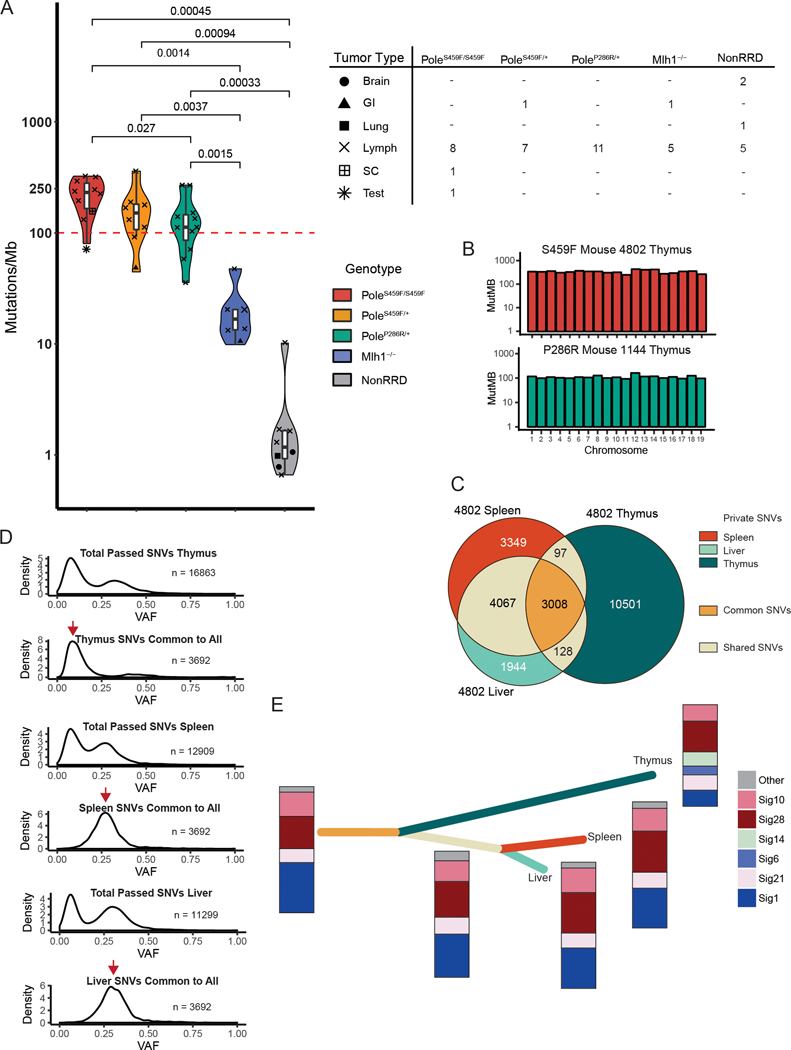
Figure 2. The genomic landscape of Pole mutant mouse tumors resembles that of POLE-driven human cancers and gives insight into mutagenesis mechanisms.
(A) Left -Violin plot showing mutation frequency from tumor whole exome sequencing (WES) of mouse tumors. Pole mutant mouse cancers (PoleS459F/S459F n = 10; PoleS459F/+ n = 8; PoleP286R/+, n = 11) were compared to MMRD mouse tumors (n=6) and replication repair proficient mouse-derived tumors (n =8). The red dashed line indicates 100 mutations/Mb. Significance are indicated using Student’s T-test. Scatter plot symbols indicate tumor type for each group and are summarized in: Right – table summarizing tumor types sequenced for each genotypic group. Brain = brain tumor; GI = gastrointestinal adenocarcinoma; Lung = lung adenocarcinoma; Lymph = lymphoma; SC = sarcoma; Test = testicular germ cell tumor.
(B) Mutation frequencies, as calculated by the number of mutations per target region covered, are plotted per chromosome, and reveal no evidence of localized hypermutation.
(C) Exome data from tumor fractions from a single mouse were compared. Private SNVs were defined as SNVs that were present only in one specific fraction. Shared SNVs were present in two fractions. Common SNVs were present in all fractions. The number of private, shared, and common SNVs are all indicated. Corresponding tumor fractions are indicated.
(D) Density plots of the number of single nucleotide variants (SNVs) by variant allele fraction (VAF) in each of the tumor fractions. SNVs unique to indicated tumor fractions and common to all tumor fractions were subsetted and plotted for each fraction. Red arrows indicate the presence of common SNVs in early and late clones for thymus vs. spleen and liver respectively.
(E) Constructed evolutionary tree for tumor 4802 based on three sequenced fractions. Length of branch is proportional to the number of SNVs. Colours correspond to private, shared, and common SNVs indicated in (C). Stacked bars indicate the proportion and presence of mutational signatures.
Mutational signatures are often indicative of specific mechanisms of mutagenesis. We plotted the proportion of tumor SNVs within specific trinucleotide contexts and performed signatures analysis using the R package deconstructSigs[39]. Signatures 10 and 28—characteristic of POLE exonuclease deficient tumors[19–21]—were found in all Pole mutant mouse tumors and together constituted the largest proportion of signatures present in each tumor (Supplementary Fig. S4).
To study how mutations accumulate in POLE mutant cancers, we examined tumors from multiple sites. As germline RRD in humans can result in multiple synchronous cancers[9], we first determined whether all lesions from several sites (liver, spleen, lymph node, and thymus) indeed originated from a single tumor. We examined sequencing data from 4 such mice (2 PoleP286R/+, 1 PoleS459F/+, and 1 PoleS459F/S459F). In all cases, there was a significant number of common SNVs present in all tumor tissues (Fig. 2C and Supplementary Fig. S5). These data suggest that a single hematopoietic malignancy arose in Pole mutant mice allowing us to study mutational processes temporally and spatially.
We next studied the mutational landscape of multi-site tumors in more detail. Despite significant overlap in mutations (>1000 SNVs shared), each tumor fraction also had a significant proportion of mutations unique to that fraction. By plotting the variant allele fractions (VAFs) of mutations unique and common to tumor fractions we were able to observe the emergence of both early and late clones (Fig. 2D and Supplementary Fig. S5), supporting a stochastic mutagenesis mechanism that acts continually.
We then used mutational analysis as described above to detect unique signatures in specific tumor clones in sites absent in other fractions. For example, PoleS459F/S459F tumors from mouse 4802 revealed signatures specific to MMR deficiency and combined MMR and POLE exonuclease deficiency only in the thymus (Fig. 2E). This was not observed in common, shared, or private SNVs in the liver and spleen. The phenomenon of complete RRD (loss of both MMR and polymerase proofreading) recapitulates our observations in human cancers which may start as polymerase mutant, and subsequently develop MMRD[15] and in cases of CMMRD that develop secondary somatic polymerase exonuclease deficiency[18].
Together, ultrahypermutation, the pattern of mutational accumulation, and signatures suggest that the underlying mutagenesis mechanism for Pol ε exonuclease deficiency is consistent between humans and mice and is a stochastic and continuous process.
Pole mutant mice develop two distinct types of T cell lymphoma
Having established the genetic processes affecting tumorigenesis in Pole mutant mice, we used high dimensional immune phenotyping to better understand whether the unique genetic mechanism affects the cellular origin and heterogeneity of lymphomas in mutant mice. We used mass cytometry to simultaneously analyze expression of 30 markers of hematopoietic cell lineage and activation/differentiation state (Supplementary Table S1) among single cells from enlarged thymus (T), spleen (S or SPL) or lymph nodes (L) of moribund PoleS459F/S459F (n=12) and PoleS459F/+(n=4) mice. Unsupervised FlowSOM clustering followed by t-Stochastic Neighbor Embedding (tSNE) revealed striking differences in the phenotype and abundance of T and B cell subsets in lymphoma samples versus those in wild-type (WT) SPL (Fig. 3A and Supplementary Fig. S6A,B). Cell clusters expressing T lineage markers (TCRβ, CD4 and/or CD8β) were more abundant in the lymphoma samples than in WT SPL, suggesting that the lymphomas were of T lineage origin. However, in some samples there were multiple T lineage clusters with varying expression of TCRβ, CD4, and/or CD8β, whereas in others there was a single predominant TCRβ+ CD4+ CD8β− cluster.
The t-SNE plots also showed that two different T cell subsets were proliferating across samples, based on incorporation of 127I-iododeoxyuridine (IdU), a thymidine analogue. There was little overlap among the T cell clusters or proliferating cells on the t-SNE plots in these two groups, suggesting two distinct T cell lymphoma subtypes, which we termed Group A and Group B. Group A lymphomas likely came from thymus where T cells develop, and subsequently tumors spread to SPL, lymph node and liver (see Supplementary Table S2). Group B lymphomas likely came from SPL or lymph node, which are secondary lymphoid tissues where naïve T and B cells undergo antigen-triggered differentiation into immune effector cells. Group B samples contained more B and myeloid cells than Group A samples (Supplementary Fig. S6B), likely reflecting their different tissues of origin. Immunohistochemistry (IHC) staining for CD3 in WT SPL, Group A and Group B tumors as well as staining for B220 in WT SPL and Group B tumors supported these findings (Supplemenary Fig. S6C).
Because non-T cells were abundant in Group B lymphomas, we generated a heatmap to better visualize group-specific expression of T cell markers among cells lacking B and myeloid cell markers. These analyses revealed significant inter-and intratumoral heterogenity both between and within Group A and Group B lymphomas. Group A samples expressed high amounts of CD8β together with variable amounts of TCRβ and CD4, but low amounts of the T cell activation markers CD44, PD1 and ICOS. Group B samples expressed high amounts of TCRβ and CD4 (with one exception) and little CD8β but expressed high amounts of T cell activation markers (Fig. 3B; Supplementary Fig. S6A). Manual analysis confirmed that Group A samples had significantly higher abundance of TCRβ+ CD8β+ and TCRβ− CD8β+ cells than Group B samples (Fig. 3A–C; Supplementary Fig. S6A). CD4 was also variably expressed by CD8β+ cells in some Group A samples (Fig. 3B, Supplementary Fig. S6A), a feature of normal immature CD4−/+ CD8+ thymocytes that proliferate extensively before differentiating into mature CD4+ and CD8+ T cells[47, 48]. Both CD8β+ subsets were proliferating in Group A samples, as indicated by their incorporation high amounts of IdU, a thymidine analogue (Fig. 3D). Thus, Group A lymphomas originate from T cell precursors, similar to human T cell lymphoblastic leukemias (also known as thymic lymphomas) that commonly arise during in mice and humans with mutations in the Ataxia telangiectasia mutated (Atm) gene or others that regulate DNA repair and DNA damage checkpoints (Supplementary Fig. S7A)[49].
By contrast, Group B lymphomas had significantly more mature TCRβ+ cells expressing CD4 but not CD8β than Group A lymphomas and WT SPL (Fig. 3A–C; Supplementary Fig. S6A). TCRβ+ cells in Group B also included significantly more CD4+ cells that co-expressed CD44, ICOS or PD1 than Group A samples or WT SPL, suggesting that they were activated (Fig. 3E; Supplementary Fig. S7B). TCRβ+ CD4+ CD8β− cells that expressed PD1 and ICOS were proliferating in Group B but not Group A or WT SPL (Fig. 3D; Supplementary Fig. S7C). Finally, Group B samples exhibited both inter- and intratumoral heterogenity with respect to activation markers (Supplementary Fig. S8A). In WT mice, these markers are expressed by antigen-activatated CD4+ T-follicular helper (TFH) cells that proliferate and differentiate in lymph nodes and spleen during immune responses to foreign antigens. These data suggest that Group B lymphomas arise in peripheral lymphoid tissues from activated TFH-like CD4 T cells.
In mice and humans, normal TFH cells promote proliferation and immunoglobulin (Ig) class switching of activated B cells in germinal centers[50]. Group B lymphomas had more class-switched IgD− IgMhi B cells expressing variable amounts of the GC B cell markers CD150, CD95 and Bcl6 than WT spleen (Supplementary Fig. 8B, C), suggesting that the TFH-like lymphoma cells acted cell non-autonomously to promote B cell proliferation and Ig class-switching in Group B tumors, as is seen in cases of human TFH-like lymphoma[51].
In Group B cases where multiple tumor-infiltrated tissues were examined (n=3), both spleen and thymus were infiltrated with TFH-like cells and exhibited similar intratumoral heterogeneity (Supplementary Fig. S8D) suggesting the same tumor migrated to different tissues. Given that Group B tumors likely originated from peripheral CD4 T cells, it is possible the lymphoma cells recirculated to the thymus as is known to occur for effector/memory T cells in normal mice.
Collectively, these data suggest that lymphomas in Pole mutant mice have distinct cells types and tissues of origin: T cell precursors in the thymus (Group A) or TFH-like T cells in peripheral lymphoid tissues (Group B).
Immune checkpoint blockade (ICB) increases proliferation in Pole mutant T cell lymphomas
Hypermutation is linked with positive responses to ICB in some solid tumors[52, 53] including RRD cancers[27, 54]. Furthermore, using immunotherapy as a preventive approach is currently discussed in the context of germline RRD carriers. We sought to determine whether prophylactic treatment with anti-PD1 and/or anti-CTLA-4 would delay lymphoma incidence and increase survival in Pole mutant mice. We focused this trial (and CyTOF analysis) on PoleS459F/S459F mice because they develop lymphoma with the shortest and least variable latency (Fig. 1A). We treated cohorts of 6-week old mutant mice twice weekly with either anti-PD1, anti-CTLA-4 or both antibodies and monitored them for tumor initiation and survival. Surprisingly, despite the extreme hypermutation, treatment with either or both checkpoint inhibitors did not significantly alter survival (Fig. 4A).
To search for a plausible explanation, we examined tumor infiltrated SPL cells from each treatment group using the same mass cytometry panel we used to characterize lymphoma in untreated mice. Heatmap visualization of T cell marker expression by cells lacking B and myeloid cell markers showed that the most abundant cells in 15/16 treated mice were TCRβ+ CD8β – cells (Fig. 4B) that resembled Group B lymphomas in the untreated cohort. Although more TCRβ+ CD8β− lymphoma cells lacked CD4 in the treated (5/15) cohort versus untreated (1/7) Group B lymphomas, they all expressed high levels CD44, PD1 and ICOS (Fig. 4B). Thus, most lymphomas in the treated cohorts were Group B tumors. We therefore asked if any of the treatments altered the abundance proliferation of activated T cells using untreated Group B samples as the comparison group. We observed no significant differences in the proportion of TCRβ+ cells or of CD4+ PD1+ and CD4+ ICOS+ cells within the TCRβ+ subset in Group B tumors from treated vs untreated mice (Fig. 4C top). These data suggest that the treatments had no impact on the abundance or phenotype of T lymphoma cells. However, the proportion of proliferating IdU+ T cells was significantly higher in mice treated with both anti-CTLA-4 and anti-PD1 (Fig. 4C bottom). This finding suggests that the combination ICB therapy failed because PD1 and CTLA-4 co-operatively restrain the proliferation of TFH-like Group B lymphoma cells that express high levels of PD1 and ICOS. These findings are consistent with previous reports on that PD1 has a tumor suppressor function in lymphoma models[55] as well as with human data from our consortium showing that human T cell lymphomas fail to respond to ICB.
DISCUSSION
In this report, we utilize novel mouse models to comprehensively study the penetrance and tumorigenic processes of POLE mutants, enabling characterization of genotype/phenotype differences observed among humans with germline POLE mutations. We find that mouse tumors driven by Pole mutations model human POLE mutant tumors genetically and have unique mechanisms of tumor progression, intra-tumoral heterogeneity, and response to ICB.
Although initially thought to be driven by MMRD alone, mutations in DNA polymerases are a major cause of RRD and hypermutation in cancer[15, 16]. Determining whether a POLE mutation can drive tumorigenesis is difficult since mutations must retain polymerase function of the protein while causing dysfunction of its proofreading ability. This large gene has been shown to harbor many mutations both in the germline and somatically in hypermutant tumors. Thus, defining true drivers is very difficult and is key for genetic counselling and initiation of surveillance protocols. While, initial reports on POLE driven mutagenesis in human cancers described a mild phenotype of adult-onset polyposis, data from our international consortium and case reports reveal that some mutations result in a more aggressive genetic syndrome (Fig. 1B). Importantly, some of the most common somatic mutations have not been reported to date in the germline. The aggressive cancer phenotype exhibited in our mouse models provides further clarity to this genotype-phenotype observation as mutations, such as P286R and S459F, may be incompatible with human development.
In humans, POLE mutations are most commonly drivers in GI and endometrial cancers[12, 16]. This is thus a limitation of studying germline PoleP286R and PoleS459F mice which develop mainly lymphomas. Nevertheless, these mutant mice are robust models to provide insight on POLE tumorigenesis as tumors recapitulate both the mutational burden and processes observed in POLE driven human tumors. Tumors from both species are driven mainly by SNVs, are evenly dispersed throughout the genome, and exhibit a similar TMB accumulation threshold. Moreover, both models exhibited specific mutations and mutational signatures consistent with POLE mutagenesis—C>A-TCT, C>T-TCG (COSMIC Signature 10)[19]; T>G-TTT (COSMIC Signature 28)[20, 21]. These signatures were not clearly observed in previous Pole-mutant models, notably in genome sequencing from LSL-PoleP286R mouse embryonic fibroblasts and tumors[31]. This may reflect differences in sequencing facility pipelines or gene dosing since LSL-PoleP286R mice are hemizygous for Pole. Differences in gene dosing and genetic backgrounds may also explain differences in tumor spectra observed.
Using several algorithms, we provide further insight on the pattern and process of Pol ε driven tumor progression. Firstly, our data suggest obligatory continuous mutation accumulation and lack of clonal stability. The constructed clonal evolutionary tree (Fig. 2E) reveals a continuous steady accumulation of mutations. This is very different than the classic clonal evolution pattern where an aggressive clone is dominant and stable, such as in melanoma[56]. Finally, it is interesting to note that both polymerase mutant and MMRD tumors can gain secondary mutations to become combined RRD in humans and mice (Fig. 2E)[18].
Human RRD hematopoietic malignancies are predominantly of T cell origin[57], but T cell malignancies arise from many different stages of T cell differentiation. Differences in cell of origin underly important differences in prognosis and outcome for many types of cancer[58]. In mice and humans, T cell lymphoblastic lymphomas arise in the thymus from TCRβ−/+ CD4−/+ CD8β+ precursors that undergo cell cycle arrest and recombine TCRA prior to completing maturation[47, 48]. Group A lymphomas in PoleS459F mutant mice were arrested during this developmental transition, which is known to be highly prone to oncogenic transformation from studies of mice and humans with genetic defects in other DNA damage/repair checkpoints[59]. Indeed, both LSL-PoleP286R mutant mice[31] as well as Pold1 proofreading deficient mice[43] also appeared to develop such lymphomas.
Outcomes for T cell lymphoblastic lymphomas are better than for mature T cell lymphomas, a heterogenous group of hematopoietic malignancies that develop when mutations accumulate during antigen-driven T cell proliferation in peripheral lymphoid tissues. This group includes angioimmunoblastic T cell lymphoma (AITL), a nodal T cell lymphoma that originates from TFH cells that harbor mutations in genes that regulate TCR signaling and epigenetic processes, occurs in older adults and has non-autonomous effects on B cells[51]. Group B lymphomas may provide a mouse model for this a rare poor prognosis type of T cell lymphoma. Collectively, our finding that PoleS459F mutant mice develop two distinct types of T cell lymphoma suggests that there are multiple stages during Pole-driven lymphomagenesis in which a T cell can gain advantage by hypermutation.
Furthermore, our detailed mass cytometry analyses reveal that, in contrast to most lymphoid malignancies, Pole driven lymphomas exhibit substantial phenotypic intratumoral heterogeneity (Fig. 3; Supplementary Fig. S8A, D). This phenotypic heterogeneity may be explained by the genomic intra-tumoral heterogeneity but also suggest that a more comprehensive analysis of human lymphomas which are driven by RRD is required to better tackle these aggressive cancers.
Treatment of PoleS459F/S459F mice prophylactically with ICB, revealed splenomegaly in all mice with clinical signs of lymphoma. Since our earlier analysis indicated that lymphoma infiltration to various organs in a single mouse was always from the same cells phenotypically (Supplementary Fig. S8D) and genomically (Fig. 2C and Supplementary Fig. S5), we therefore focused our analysis of tumor-infiltrated spleen cells from all treatment groups. Upon analysis by CyTOF, we observed that 15/16 treated tumors were Group B with 1/16 classifying as Group A (Fig. 4B). This enrichment is important since PD1 was not highly expressed by mouse Group A lymphomas and this can explain the survival benefit for Group B tumors and their lack of response to (and potentially exacerbation by) ICB. In hypermutant solid tumors ICB is used to counteract the PD1 immune inhibitory checkpoint restraints, unleashing intra-tumoral CD8 T cells to kill tumor cells that express mutationally generated “neoantigens”[53, 60]. However, prophylactic treatment of Pole mutant mice with anti-PD1 alone or in combination with anti-CTLA-4 did not improve survival or decrease the abundance of TFH-like CD4 cells.
While this lack of therapeutic ICB effect could reflect “immuno-editing” or other upregulation of other inhibitory checkpoints[52, 61, 62], it more likely reflects the unique biology of the Group B lymphomas, which had few CD8 T cells and consisted predominantly of TFH-like CD4 cells expressing high levels of PD1, ICOS and CD44. PD1[50, 63] and CTLA-4[64] can both restrain ICOS-induced co-stimulation of normal TFH cells. Thus, combined interference with the PD1 and CTLA-4 inhibitory checkpoints on malignant Group B T cell lymphomas may allow ICOS-mediated co-stimulation to promote their growth. In accordance with this idea, proliferation of the TFH-like lymphoma was increased by combination ICB therapy (Fig. 4D). This finding accords with a recent study showing that anti-CTLA-4 expands ICOS+ TH1-like CD4 T cells in murine tumor models[65]. Interestingly, mono- and bi-allelic deletions of PDCD1, the gene encoding PD1, are frequent in some human T cell lymphoma types[55], suggesting a tumor suppressor function in these cases. While these data may not be generalizable to human solid tumors driven by POLE mutations (indeed ICB has been shown to effectively treat hypermutant solid tumors[27, 54]), this tumor suppressor function of PD1 may explain data from our international consortium suggest that anti-PD1 therapy will not prevent or delay occurrences of T cell malignancies in RRD mutant patients. Taken together, our findings suggest that the use of ICB therapies warrants urgent further investigation for PD1+ T cell lymphomas or RRD hematological malignancies.
In summary, our mouse models provide a platform to study hypermutant lymphomas. They provide valuable information about specific mutations and their potential phenotype in humans. Establishing the kinetics and clonal evolution of RRD cancers, which cannot be done by studying yeast or human tumors, may be used as an Achilles’ heel for these cancers. Finally, this highly penetrant model of RRD tumors may be the most reliable approach for future robust preclinical testing of immune-based drugs on spontaneous tumors rather than established cell lines on immunocompromised mice.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Two mouse models of polymerase exonuclease deficiency shed light on mechanisms of mutation accumulation and considerations for immunotherapy.
ACKNOWLEDGEMENTS:
We thank Lauryl Nutter and Marina Gertsenstein (TCP Transgenic Core) and TCP staff for their scientific advice and technical support. The Tabori lab research is supported by a Stand Up To Cancer- Bristol-Myers Squibb Catalyst Research Grant (Grant Number: SU2C-AACR-CT-07–17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research Grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. The Tabori lab is also supported by Meagan’s Walk (MW-2014–10), b.r.a.i.n.child Canada, LivWise, a Canadian Institutes for Health Research (CIHR) Grant (PJT-156006), and the CIHR Joint Canada-Israel Health Research Program. The Pursell and Suo labs have been funded by the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH) under award number (R01ES028271). The Pursell lab has also been funded by a Tulane University Carol Lavin-Bernick Faculty Grant. The Guidos lab acknowledges support from the Ontario Institute for Cancer Research with funding from the Government and Ontario, as well as the Garron Family Cancer Center of the Hospital for Sick Children Research Institute. M.A.G is supported by the Government of Ontario, The Garron Family Cancer Centre, and the Peterborough K.M. Hunter Graduate Program.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have declared no conflicts of interest
REFERENCES
- 1.Drake JW, et al. , Rates of spontaneous mutation. Genetics, 1998. 148(4): p. 1667–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K and Kunkel TA, Functions of DNA polymerases. Adv Protein Chem, 2004. 69: p. 137–65. [DOI] [PubMed] [Google Scholar]
- 3.Hogg M, et al. , Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol, 2014. 21(1): p. 49–55. [DOI] [PubMed] [Google Scholar]
- 4.Lange SS, Takata K, and Wood RD, DNA polymerases and cancer. Nat Rev Cancer, 2011. 11(2): p. 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyabe I, Kunkel TA, and Carr AM, The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet, 2011. 7(12): p. e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pursell ZF, et al. , Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science, 2007. 317(5834): p. 127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganai Rais A. and Johansson E, DNA Replication—A Matter of Fidelity. Molecular Cell, 2016. 62(5): p. 745–755. [DOI] [PubMed] [Google Scholar]
- 8.Haraldsdottir S, et al. , Colon and Endometrial Cancers With Mismatch Repair Deficiency Can Arise From Somatic, Rather Than Germline, Mutations. Gastroenterology, 2014. 147(6): p. 1308–1316.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakry D, et al. , Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer, 2014. 50(5): p. 987–96. [DOI] [PubMed] [Google Scholar]
- 10.Lynch HT and de la Chapelle A, Hereditary colorectal cancer. N Engl J Med, 2003. 348(10): p. 919–32. [DOI] [PubMed] [Google Scholar]
- 11.Palles C, et al. , Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet, 2013. 45(2): p. 136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Church DN, et al. , DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet, 2013. 22(14): p. 2820–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Cancer Genome Atlas, N., et al. , Comprehensive molecular characterization of human colon and rectal cancer. Nature, 2012. 487: p. 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine DA, et al. , Integrated genomic characterization of endometrial carcinoma. Nature, 2013. 497: p. 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell BB, et al. , Comprehensive Analysis of Hypermutation in Human Cancer. Cell, 2017. 171(5): p. 1042–1056.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briggs S and Tomlinson I, Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol, 2013. 230(2): p. 148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johanns TM, et al. , Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discovery, 2016. 6(11): p. 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlien A, et al. , Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet, 2015. 47(3): p. 257–62. [DOI] [PubMed] [Google Scholar]
- 19.Alexandrov LB, et al. , Signatures of mutational processes in human cancer. Nature, 2013. 500(7463): p. 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov LB, et al. , The Repertoire of Mutational Signatures in Human Cancer. bioRxiv, 2018: p. 322859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petljak M, et al. , Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell, 2019. 176(6): p. 1282–1294.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbari SR, et al. , Functional Analysis of Cancer-Associated DNA Polymerase ε Variants in Saccharomyces cerevisiae. G3: Genes|Genomes|Genetics, 2018. 8(3): p. 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinbrot E, et al. , Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res, 2014. 24(11): p. 1740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodel KP, et al. , Explosive mutation accumulation triggered by heterozygous human Pol epsilon proofreading-deficiency is driven by suppression of mismatch repair. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder A, et al. , Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med, 2014. 371(23): p. 2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Allen EM, et al. , Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 2015. 350(6257): p. 207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le DT, et al. , PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med, 2015. 372(26): p. 2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizvi NA, et al. , Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albertson TM, et al. , DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A, 2009. 106(40): p. 17101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korona DA, Lecompte KG, and Pursell ZF, The high fidelity and unique error signature of human DNA polymerase epsilon. Nucleic Acids Res, 2011. 39(5): p. 1763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HD, et al. , Polymerase-mediated ultramutagenesis in mice produces diverse cancers with high mutational load. J Clin Invest, 2018. 128(9): p. 4179–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rayner E, et al. , A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer, 2016. 16(2): p. 71–81. [DOI] [PubMed] [Google Scholar]
- 33.Barbari SR and Shcherbakova PV, Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy. DNA Repair (Amst), 2017. 56: p. 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gertsenstein M and Nutter LMJ, Engineering Point Mutant and Epitope-Tagged Alleles in Mice Using Cas9 RNA-Guided Nuclease. Current Protocols in Mouse Biology, 2018. 8(1): p. 28–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edelmann W, et al. , Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res, 1999. 59(6): p. 1301–7. [PubMed] [Google Scholar]
- 36.Zahurancik WJ, et al. , Comparison of the kinetic parameters of the truncated catalytic subunit and holoenzyme of human DNA polymerase varepsilon. DNA Repair (Amst), 2015. 29: p. 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahurancik WJ, Klein SJ, and Suo Z, Significant contribution of the 3’-->5’ exonuclease activity to the high fidelity of nucleotide incorporation catalyzed by human DNA polymerase. Nucleic Acids Res, 2014. 42(22): p. 13853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danecek P, et al. , The variant call format and VCFtools. Bioinformatics, 2011. 27(15): p. 2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenthal R, et al. , deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biology, 2016. 17(1): p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An Y, et al. , Cdh1 and Pik3ca Mutations Cooperate to Induce Immune-Related Invasive Lobular Carcinoma of the Breast. Cell Rep, 2018. 25(3): p. 702–714.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Gassen S, et al. , FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A, 2015. 87(7): p. 636–45. [DOI] [PubMed] [Google Scholar]
- 42.Amir el AD, et al. , viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol, 2013. 31(6): p. 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldsby RE, et al. , High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc Natl Acad Sci U S A, 2002. 99(24): p. 15560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reitmair AH, et al. , MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet, 1995. 11(1): p. 64–70. [DOI] [PubMed] [Google Scholar]
- 45.Prolla TA, et al. , Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet, 1998. 18(3): p. 276–9. [DOI] [PubMed] [Google Scholar]
- 46.Xing X, et al. , A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat Commun, 2019. 10(1): p. 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guidos CJ, Weissman IL, and Adkins B, Intrathymic maturation of murine T lymphocytes from CD8+ precursors. Proc Natl Acad Sci U S A, 1989. 86(19): p. 7542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boudil A, et al. , IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection. Nat Immunol, 2015. 16(4): p. 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matei IR, Guidos CJ, and Danska JS, ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: the DSB connection. Immunol Rev, 2006. 209: p. 142–58. [DOI] [PubMed] [Google Scholar]
- 50.Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity, 2019. 50(5): p. 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallois D, et al. , Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood, 2016. 128(11): p. 1490–502. [DOI] [PubMed] [Google Scholar]
- 52.Rooney MS, et al. , Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell, 2015. 160(1–2): p. 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGranahan N, et al. , Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science, 2016. 351(6280): p. 1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouffet E, et al. , Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. Journal of Clinical Oncology, 2016. [DOI] [PubMed] [Google Scholar]
- 55.Wartewig T, et al. , PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birkeland E, et al. , Patterns of genomic evolution in advanced melanoma. Nature Communications, 2018. 9(1): p. 2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wimmer K, et al. , Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘Care for CMMRD’ (C4CMMRD). Journal of Medical Genetics, 2014. 51(6): p. 355–365. [DOI] [PubMed] [Google Scholar]
- 58.Visvader JE, Cells of origin in cancer. Nature, 2011. 469(7330): p. 314–22. [DOI] [PubMed] [Google Scholar]
- 59.Danska JS, et al. , Rescue of T cell-specific V(D)J recombination in SCID mice by DNA-damaging agents. Science, 1994. 266(5184): p. 450–5. [DOI] [PubMed] [Google Scholar]
- 60.Sharma P and Allison JP, Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell, 2015. 161(2): p. 205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma P, et al. , Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell, 2017. 168(4): p. 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zappasodi R, Merghoub T, and Wolchok JD, Emerging Concepts for Immune Checkpoint Blockade-Based Combination Therapies. Cancer Cell, 2018. 34(4): p. 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi J, et al. , PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity, 2018. 49(2): p. 264–274.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sage PT, et al. , The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity, 2014. 41(6): p. 1026–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei SC, et al. , Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell, 2017. 170(6): p. 1120–1133.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during our study have been deposited in the NCBI’s Sequence Read Archive (SRA) database under the accession code PRJNA659565.