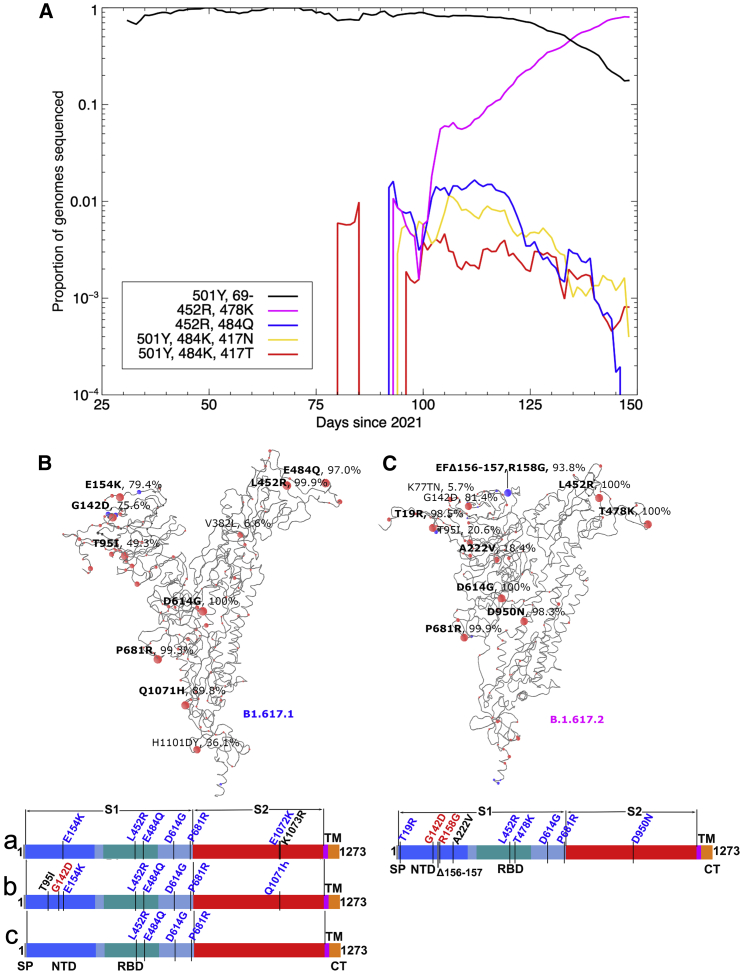
Figure 1.
Mutational landscape of the B.1.617 lineage
(A) Evolution plot showing trajectories of various mutations in the COG-UK data. Certain mutations were used to select for sequences typically belonging to a given strain: 501Y and Δ69 to select the B.1.1.7 variant; 501Y, 484K, and 417N to select the B.1.351 variant; 501Y, 484K, and 417T to select the P.1 variant; E484Q and L452R to select the B.1.617.1 variant; and T478K and L452R to select the B.1.617.2 variant.
(B and C) Schematic showing the locations of amino acid substitutions in B.1.617.1 (B) and B.1.617.2 (C) relative to the ChAdOx1 SARS-CoV-2 sequence, as drawn in previous studies (Dejnirattisai et al., 2021a, 2021b; Supasa et al., 2021; Zhou et al., 2021), with all amino acid mutations above 5% explicitly labeled. Mutations colored in bold were included in the constructs used in this study for the given strain. Under the structural cartoon is a linear representation of S with changes marked for B.1.617.2 live virus, and the three subvariants of B.1.617.1 (a, b, and c) used in this study are detailed. Where there is a charge change introduced by mutations, the change is colored (red when the change makes the mutant more acidic/less basic and blue for more basic/less acidic).