Figure 1:
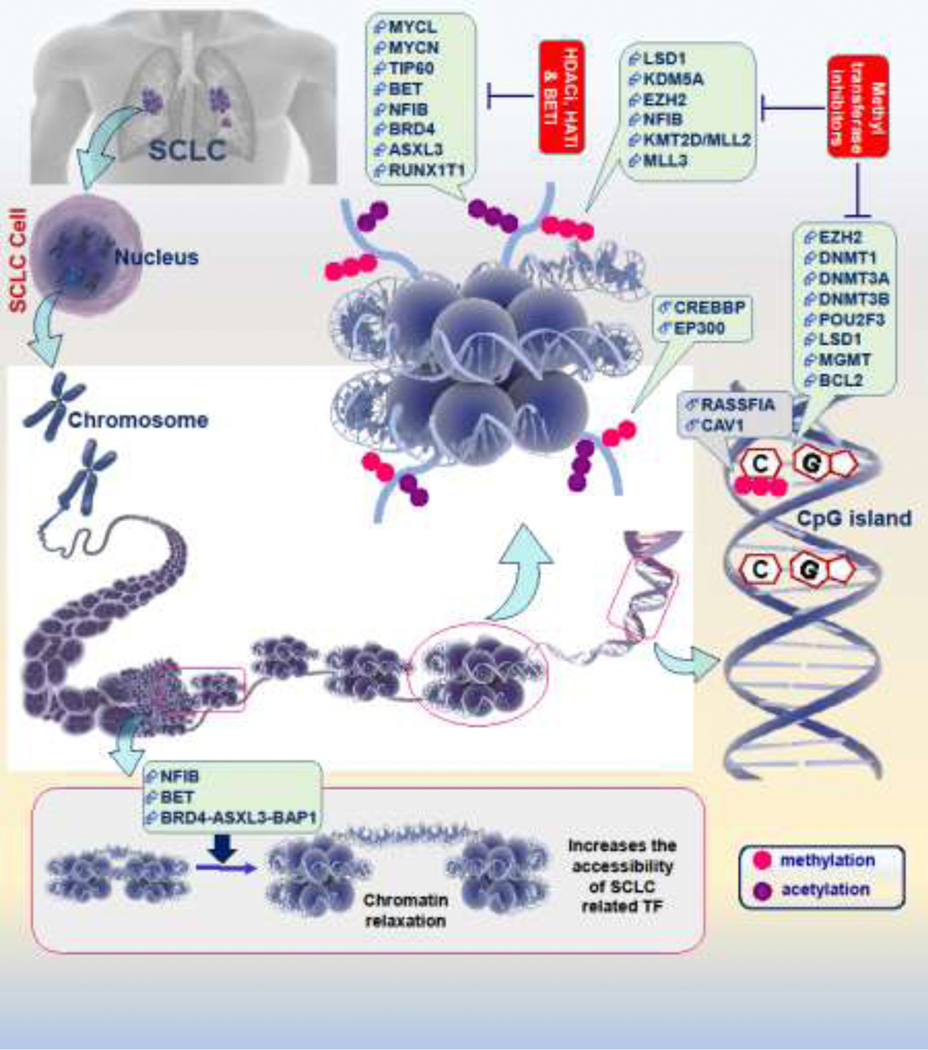
Epigenetic landscape of SCLC genome is regulated at the DNA & histone level by multiple regulators. The prime features of this regulation involve modifications of basic chromatin unit ‘nucleosome’ an octamer of histone that works as a spool for DNA wrapping. The key post-translational modifications of histones mainly involves acetylation and methylation of lysine residues that controls chromatin transformations and regulates the expression of target genes (other modifications like sumoylation, phosphorylation, and ubiquitination, are not shown here). A number of chromatin modifier proteins are involved performing this task including readers, writers, and erasers (like HDACS, HATs, and methyltransferases). SCLC-specific key epigenetic genes that regulate the histone/DNA related epigenetic modifications have been reviewed and summarized in the illustration. Similar to writer, readers, and erasers, the overexpression of other factors like NFIB induces the chromatin relaxation (as shown in the lower side panel) and increases the accessibility of transcription factors to the DNA that ultimately induces the expression of SCLC related genes. Epigenetic drugs (HDACi: histone deacetylase inhibitors, HATi: histone acetyltransferase inhibitors, BETi: bromodomain and extra-terminal domain protein inhibitors, and inhibitors of methyltransferases) targets these chromatin modifiers.
