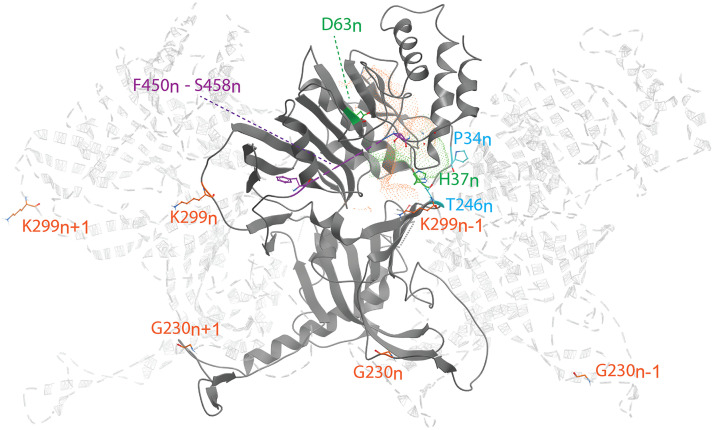
FIG 6.
Key amino acid residues linked to resistance to CHIKV nsP1 inhibitors, mapped to the CHIKV nsP1 cryo-EM structure (PDB code 6Z0V), represented as three consecutive nsP1 protomers: n, n+1, and n–1. Residues are color-coded as follows: green, catalytic residues H37 and D63; blue, residues P34 and T246, involved in resistance to the MADTP series; orange, residues G230 and K299, conferring resistance to the FHNA series. Residues F450 to S458 (purple) flank the area where residues S454 and W456, involved in resistance to the CHVB series, would be located. Orange and green dotted volumes represent the proposed binding pockets of GTP and SAM, respectively, for reference.