Abstract
Sparganium fallax is an aquatic perennial herb distributed in eastern Asia. The complete chloroplast genome of S. fallax was sequenced and assembled. The genome size was 161,838 bp in length with 36.8% GC content. Its quadripartite structure consisted of the large single copy (LSC, 89,042 bp) and small single copy (SSC, 18,774bp) regions, separated by a pair of inverted repeats (IRS) of 27,011bp. The genome contained 114 genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis within order Poales including three Sparganium species showed that Sparganium is monophyletic and most closely related to Typha.
Keywords: Chloroplast genome, phylogenetic analysis, Sparganium fallax, Typhaceae
Sparganium L. is an aquatic perennial genus including approximately 14-19 species (Cook and Nicholls 1986, 1987; Kaul 2000; Sun and Simpson 2010). It occurs primarily in temperate and cool regions and is ecologically important in aquatic communities (Sulman et al. 2013). To date, the complete chloroplast (cp) genomes of two Sparganium species, S. eurycarpum subsp. coreanum (H.Lév.) C.D.K.Cook & M.S.Nicholls and S. stoloniferum (Buch.-Ham. ex Graebn.) Buch.-Ham. ex Juz. have been reported (Gil et al. 2019; Su et al. 2019). These two species composed one of the two clades of Sparganium (Sulman et al. 2013). The cp genome data of species from the other clade of Sparganium have not been reported. Sparganium fallax Graebn. is an important dominant species in highland wetlands in eastern Asian (Sun and Simpson 2010) and placed in the other clade of Sparganium (Sulman et al. 2013). Here, we reported complete cp genome of S. fallax and reconstructed phylogenetic tree for intention of confirming the monophyly of Sparganium.
The sample of S. fallax was collected from Jianning, Fujian, China (116.89°E, 26.81°N). A specimen was deposited at the herbarium of Wuhan University (www.whu.edu.cn, Xinwei Xu, xuxw@whu.edu.cn) under the voucher number Xu3372. Total genomic DNA was extracted using the DNA Secure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol. Library preparation and genomic sequencing on the Illumina Hiseq 2500 platform were conducted by Benagen (Benagen Inc., Wuhan, China). The obtained paired-end reads were assembled using SPAdes v.3.9.0 (Nurk et al. 2017). The assembled sequence was annotated in MPI-MP CHLOROBOX (https://chlorobox.mpimp-golm.mpg.de/geseq.html) via GeSeq (Tillich et al. 2017) with the reference cp genome of Typha latifolia L. (Guisinger et al. 2010; NC_013823.1), and then corrected using Geneious Prime v2020.2 (Kearse et al. 2012). Finally, the complete chloroplast genome of S. fallax was submitted to GenBank (Accession No. MW583115).
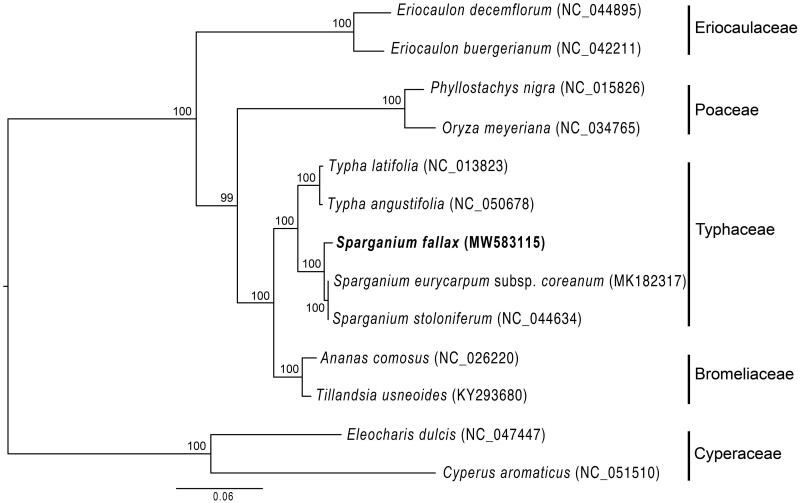
The chloroplast genome of S. fallax was 161,838 bp in length, including a large single copy (LSC) region of 89,042, a small single copy (SSC) region of 18,774 bp, and two inverted repeat (IR) regions of 27,011bp. The GC contents of LSC, SSC, IR and whole genome are 34.7%, 30.7%, 42.4%, and 36.8%, respectively. There are 114 genes annotated, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Of the 18 genes containing introns, three of them have two introns (ycf3, clpP, and rps12). Maximum likelihood analysis of Poales was conducted based on cp genomes of 13 species using RAxML v.8 (Stamatakis 2014) with 1000 bootstrap (BS) replications. The phylogenetic tree confirmed the monophyly of Sparganium with strong support (100% BS), which consisted of two clades including S. fallax and S. eurycarpum subsp. coreanum plus S. stoloniferum, respectively, and was most closely related to Typha (Figure 1).
Figure 1.
The maximum likelihood tree inferred from 13 representative chloroplast genomes of order Poales. The position of Sparganium fallax is highlighted in bold and numbers above each node are bootstrap support values.
Funding Statement
This work was supported by the Special Project of Basic Work of Science and Technology, Ministry of Science and Technology, China under Grant Number [2013FY112300].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW583115. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA703417, SRR13684638, and SAMN17861312, respectively.
References
- Cook CDK, Nicholls MS.. 1986. A monographic study of the genus Sparganium (Sparganiaceae). Part 1, Subgenus Xanthosparganium Holmberg. Bot Helv. 96:213–267. [Google Scholar]
- Cook CDK, Nicholls MS.. 1987. A monographic study of the genus Sparganium (Sparganiaceae). Part 2, Subgenus Sparganium. Bot Helv. 97:1–44. [Google Scholar]
- Gil HY, Ha YH, Choi KS, Lee JS, Chang KS, Choi K.. 2019. The chloroplast genome sequence of an aquatic plant, Sparganium eurycarpum subsp. coreanum (Typhaceae). Mitochondrial DNA Part B. 4(1):684–685. [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK.. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in poaceae. J Mol Evol. 70(2):149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul RB, Flora of North America Editorial Committee 2000. Sparganiaceae. In: Flora of North America North of Mexico. Vol. 22: Magnoliophyta: Alismatidae, Arecidae, Commelinidae (in Part) and Zingiberidae. New York and Oxford: Oxford University Press; p. 270–277. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeyniko A, Pevzner PA.. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Yang JX, Lin YG, Kang N, Hu GX.. 2019. Characterization of the complete chloroplast genome of Sparganium stoloniferum (Poales: Typhaceae) and phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1402–1403. [Google Scholar]
- Sulman JD, Drew BT, Drummond C, Hayasaka E, Sytsma KJ.. 2013. Systematics, biogeography, and character evolution of Sparganium (Typhaceae): diversification of a widespread, aquatic lineage. Am J Bot. 100(10):2023–2039. [DOI] [PubMed] [Google Scholar]
- Sun K, Simpson DA.. 2010. Typhaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 23 Acoraceae through Cyperaceae. Beijing (China): Science Press; St, Louis (MI): Missouri Botanical Garden Press; p. 158–163. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW583115. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA703417, SRR13684638, and SAMN17861312, respectively.