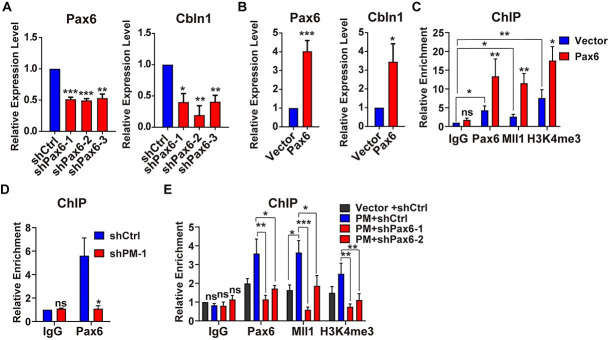
Fig 5. Pax6 is required for the lncRNA-PM-mediated Mll1-H3K4me3 recruitment.
(A) The expression levels of Pax6 and Cbln1 mRNAs in the control or Pax6 shRNAs-treated Neuro2a cells. All results were normalized to Gapdh. Data are shown as means ± SEMs, n = 3. (B) The expression levels of Pax6 and Cbln1 mRNAs in the control (Vector) or Pax6-overexpressed Neuro2a cells. All results were normalized to Gapdh. Data are shown as means ± SEMs, n = 3. (C) ChIP-qPCR detection of the indicated Pax6, Mll1, and H3K4me3 recruitment on the 5′ regulatory region of Cbln1 in the control or Pax6-overexpressed Neuro2a cells. Data are shown as means ± SEMs, n = 3. (D) ChIP-qPCR detection of Pax6 on the 5′ regulatory region of Cbln1 in the control or the indicated shPM in Neuro2a cells. Data are shown as means ± SEMs, n = 3. (E) Changes of the Pax6, Mll1, and H3K4me3 deposition on the 5′ regulatory region of Cbln1 in PM-overexpressed Neuro2a cells, with the treatments of Pax6 (shPax6-1 and shPax6-2) or control shRNAs (shCtrl). All the data of this figure can be found in the S1 Data file. Data are shown as means ± SEMs, n = 3. *P < 0.05, **P < 0.01, and ***P < 0.001. Cbln1, Cerebellin-1; ChIP-qPCR, chromatin immunoprecipitation couple with quantitative PCR; IgG, immunoglobulin G; lncRNA-PM, lncRNA-Promoting Methylation; ns, no significance.