Fig 6. In vivo association of lncRNA-PM and Pax6/Mll1 complex with Cbln1.
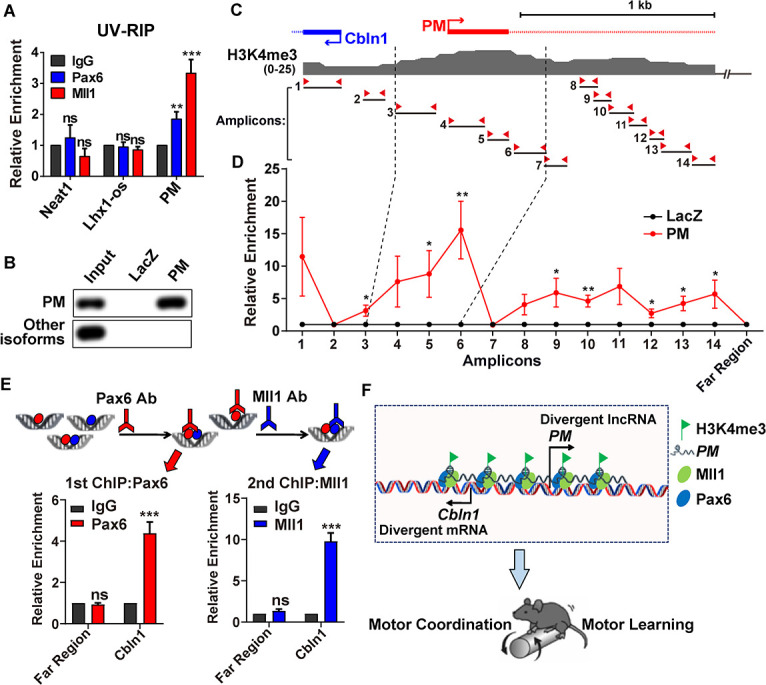
(A) UV-RIP assay showed PM:Pax6 and PM:Mll1 interactions in mouse cerebellum. Neat1 and lncRNA-lhx1os: negative controls. Data are shown as means ± SEMs, n = 3. (B) RT-qPCR detection following PM RNA pull-down using cerebellum tissue. PM: PM probes; LacZ: control probes. (C) Illustration of H3K4me3 signature on the upstream regulatory region of Cbln1 locus from +300 to −2,000 base pair based on ENCODE data in mouse cerebellum. The 14 pairs of amplicons are indicated. (D) ChIRP-qPCR showed occupancy of PM to the regions amplified by the indicated 14 amplicons and a control region that is 85 kb upstream of Cbln1 transcription start site (Far Region). Data are shown as means ± SEMs, n = 3. (E) Re-ChIP of Pax6 and Mll1. Top: illustrated flowchart of double ChIP experiment. Bottom left: recruitments of Pax6 or IgG to Cbln1 or the negative control region (Far Region) after first ChIP in mouse cerebellum. Bottom right: recruitments of Mll1 or IgG to Cbln1 or Far Region after second ChIP in mouse cerebellum. Data are shown as means ± SEMs, n = 3. (F) Model: LncRNA-PM mediates the transcriptional activation of Cbln1 through Pax6/Mll1-mediated H3K4me3 and maintains mouse motor coordination and motor learning. All the data of this figure can be found in the S1 and S2 Data files. *P < 0.05, **P < 0.01, and ***P < 0.001. Cbln1, Cerebellin-1; ChIP, chromatin immunoprecipitation; ChIRP, chromatin isolation by RNA purification; IgG, immunoglobulin G; lncRNA-PM, lncRNA-Promoting Methylation; ns, no significance; qPCR, quantitative PCR; Re-ChIP, Re-chromatin immunoprecipitation; RT-qPCR, reverse transcription-quantitative PCR; UV-RIP, UV-RNA immunoprecipitation.
