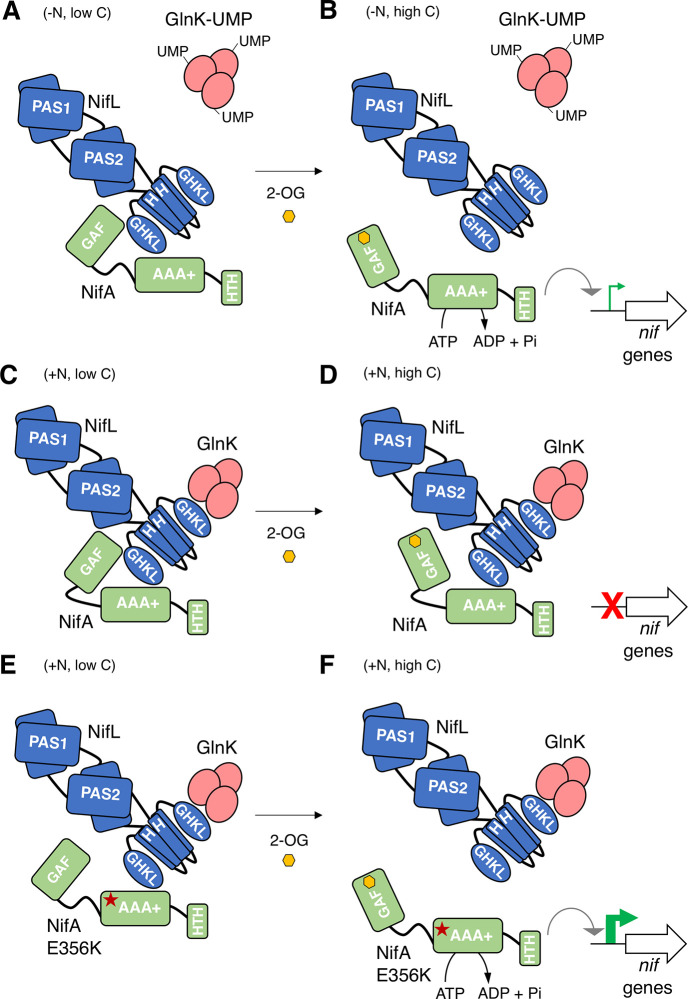
Fig 6. Model for 2-oxoglutarate regulation of NifA activity based on genetic and biochemical experiments.
(A) When 2-oxoglutarate levels are low, NifL can inhibit NifA even under nitrogen-limiting conditions (-N, low C) when GlnK is uridylylated (GlnK-UMP) and unable to interact with NifL. (B) Binding of 2-oxoglutarate (2-OG, yellow hexagon) to the GAF domain of NifA under carbon sufficient conditions (-N, high C), disrupts the binary NifL-NifA interaction, thus activating NifA. (C) Under nitrogen excess conditions (+N, low C), non-covalently modified GlnK, interacts with the GHKL domain of NifL, stimulating the formation of a ternary complex between GlnK, NifL and NifA that inhibits NifA activity. (D) The GlnK-NifL-NifA ternary complex is stable under nitrogen excess conditions even if the GAF domain in NifA is saturated with 2-OG (+N, high C). (E) The E356K substitution in the AAA+ domain of NifA (red star), perturbs the interaction with NifL in the absence of the non-modified form of GlnK. However, under nitrogen excess conditions, the GlnK-NifL-NifA-E356K ternary complex is formed when 2-OG is limiting, as a consequence of carbon limitation (+N, low C). (F) Upon a switch to a preferred carbon source, when 2-OG levels are sufficient (+N, high C), conformational changes triggered by 2-OG binding to the GAF domain disrupt the ternary complex, activating NifA-E356K under conditions of nitrogen excess.