Figure 1. General scheme for the isolation of active sequences from in vitro evolution experiments, HTS and the analysis of the sequencing data.
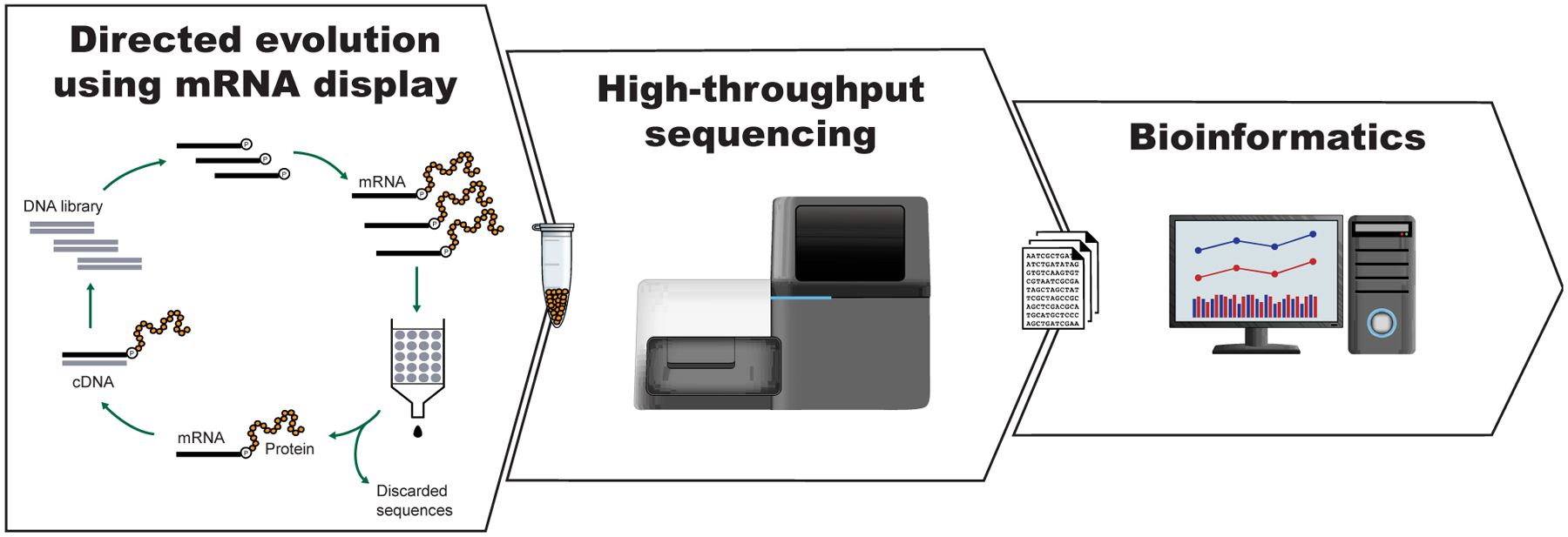
A large library of mutant variants is subjected to a selection process in which survival depends on the ability to carry out a specific biochemical function (e.g., binding). Selected variants are isolated and amplified while unselected variants are discarded. The cycle of selection and amplification is repeated several times (rounds) until variants with high activity dominate the library. The final library (and possibly intermediate pools) is then sequenced, such as by using high throughput sequencing (HTS) technologies. Finally, HTS data is analyzed using bioinformatic tools appropriate for the project’s goal. For a more detailed explanation of the selection process see Figure 2.
