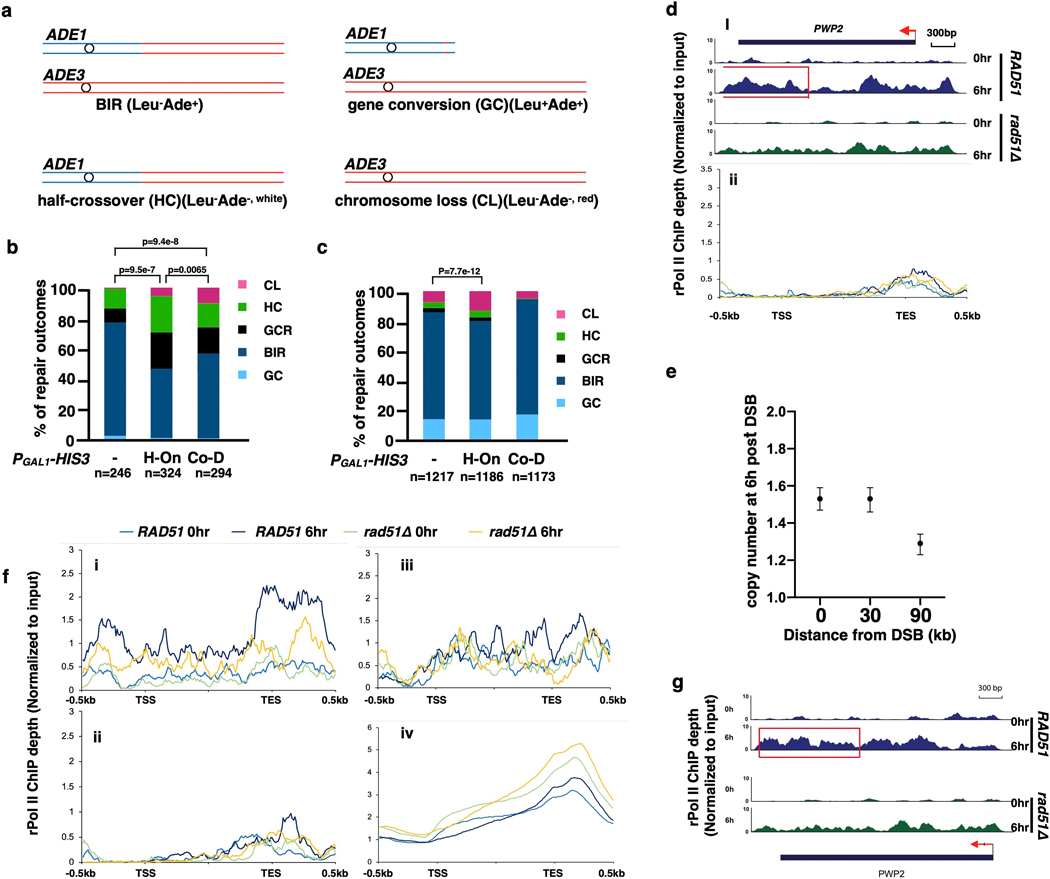
Extended Data Figure 9. BIR is interrupted by transcription.
a, Products of DSB repair as distinguished by genetic markers. b, Distribution of DSB repair outcomes for strains with and without PGAL1-HIS3 inserted at MATα-inc. c, the same as in b, but for strains with PGAL1-HIS3 inserted at 6kb position. In b and c, sample sizes (from 3 biological repeats) are indicated (see Methods for details). Statistical comparisons are performed using Chi-square test (two-sided, df=1); p values are indicated. d, (i) rPolII enrichment at TES detected by ChIP-seq for PWP2 gene located 20kb centromere distal to MAT. Red rectangle: BIR-promoted accumulation of rPolII at TES; (ii) rPolII distribution for all H-On genes located on donor chromosome III between 60 and 90 kb centromere distal to MAT. Data from the same experiment as shown in Fig. 4f. e, AMBER analysis of BIR synthesis from the same samples as in Fig. 4f (n=1, mean values of ratios between target and reference (ACT1) loci are calculated using Poisson distribution based on 10,000 droplets with error bars representing upper and lower Poisson 95%CI.). f, independent repeat of experiment shown in Fig. 4f. Metagene plot showing the distribution of normalized rPolII-ChIP-seq reads along chromosome III. i) At genes located within 0–30kb centromere distal of the MAT locus (n=10) in H-On direction; ii) at genes located within 60–90kb centromere distal to the MAT locus in H-On orientation (n=11); iii) at genes located within 0–30kb centromere distal to the MAT locus in Co-D orientation (n=9); iv) at all the genes except genes on chromosome III (n=2950). Transcription start site (TSS), transcription end site (TES) and 500 bp flanking regions were plotted. Y axis showing the mean value of rPolII depth. g, Example of BIR-caused rPolII re-distribution in yeast gene PWP2 located on chrIII (data from same experiment in Extended Data Fig. 9f). The panels show normalized rPolII ChIP-seq reads from RAD51 (wild type) (top) and rad51Δ (bottom). Red rectangle: BIR-promoted rPolII accumulation.