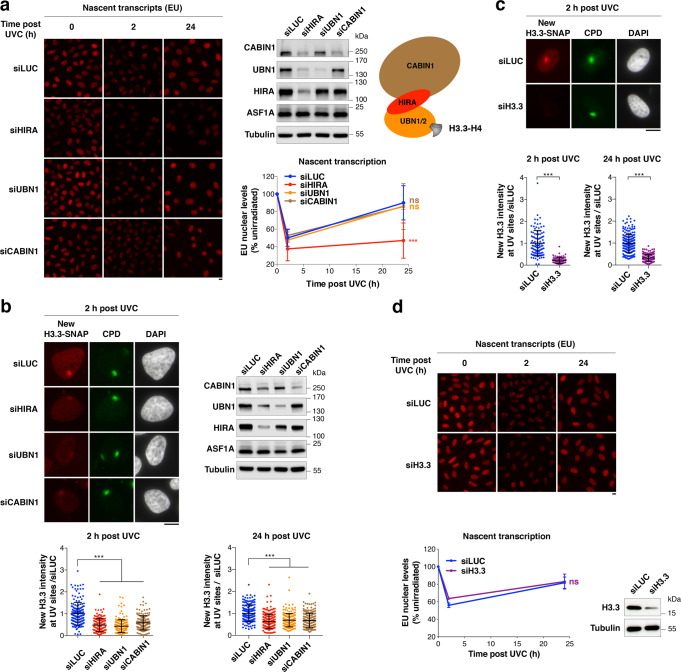
Fig. 1. HIRA-mediated transcription restart post UVC damage is independent of new H3.3 deposition.
a Fluorescence images showing nascent transcripts labelled with ethynyl-uridine (EU) in unirradiated (0 h) or UVC-irradiated (2 h, 24 h) HeLa cells treated with the indicated siRNAs (siLUC, control). Knock-down efficiencies are controlled by western blot at the time of UV irradiation (Tubulin, loading control). The scheme on the right represents the HIRA complex chaperoning H3.3-H4. The graph shows quantification of nascent transcript levels post UVC relative to unirradiated cells using the same colour code as the scheme for HIRA complex subunits. b, c New H3.3 accumulation at UVC damage sites (marked by CPD immunostaining) analysed 2 h and 24 h post local UVC irradiation in U2OS H3.3-SNAP cells treated with the indicated siRNAs (siLUC, control). The scatter plots show new H3.3 levels at UV sites (mean ± s.d. from at least 86 cells scored in three independent experiments). siRNA efficiencies are controlled by western blot for HIRA complex subunits (Tubulin, loading control), and by new H3.3-SNAP labelling for H3.3. d Nascent transcript levels analysed by ethynyl-uridine incorporation (EU) as in (a). The line graphs show mean values ± s.d. from four (a) and three (d) independent experiments scoring at least 81 cells per experiment. Statistical significance is calculated by two-way (a, d) and one-way ANOVA (b) with Bonferroni posttest, and via two-sided Student’s t-test with Welch’s correction (c). ***: p < 0.001; ns: p > 0.05. Scale bars, 10 μm. Source data are provided as a Source Data file.