Abstract
Objectives
Non‐sputum‐based tests to accurately identify active tuberculosis (TB) disease and monitor response to therapy are urgently needed. This study examined the biomarker capacity of a panel of plasma proteins alone, and in conjunction with a previously identified miRNA signature, to identify active TB disease.
Methods
The expression of nine proteins (IP‐10, MCP‐1, sTNFR1, RANTES, VEGF, IL‐6, IL‐10, TNF and Eotaxin) was measured in the plasma of 100 control subjects and 100 TB patients, at diagnosis (treatment naïve) and over the course of treatment (1‐, 2‐ and 6‐month intervals). The diagnostic performance of the nine proteins alone, and with the miRNA, was assessed.
Results
Six proteins were significantly up‐regulated in the plasma of TB patients at diagnosis compared to controls. Receiver operator characteristic curve analysis demonstrated that IP‐10 with an AUC = 0.874, sensitivity of 75% and specificity of 87% was the best single biomarker candidate to distinguish TB patients from controls. IP‐10 and IL‐6 levels fell significantly within one month of commencing treatment and may have potential as indicators of a positive response to therapy. The combined protein and miRNA panel gave an AUC of 1.00. A smaller panel of only five analytes (IP‐10, miR‐29a, miR‐146a, miR‐99b and miR‐221) showed an AUC = 0.995, sensitivity of 96% and specificity of 97%.
Conclusions
A novel combination of miRNA and proteins significantly improves the sensitivity and specificity as a biosignature over single biomarker candidates and may be useful for the development of a non‐sputum test to aid the diagnosis of active TB disease.
Keywords: diagnosis, microRNA, plasma biomarkers, proteins, tuberculosis
The biomarker potential of plasma proteins to identify tuberculosis (TB) patients was examined. Six proteins were significantly elevated in TB patients with IP‐10 and IL‐6 declining with treatment. In conjunction with miRNA previously detected, a five analyte biosignature could identify TB patients with an AUC = 0.995, sensitivity of 96% and specificity of 97%. IP‐10 was the best single biomarker candidate, showing utility as a triage tool, to quickly and easily identify potential TB patients and monitor their response to therapy.

Introduction
Tuberculosis (TB) is the leading cause of death from an infectious agent globally, with 10 million new cases in 2019, of which an estimated three million were undiagnosed and therefore did not receive treatment. 1 Detecting new cases of TB disease is usually a passive process, when individuals who have symptoms seek medical care. A recent cluster randomised control trial by Marks et al. 2 demonstrated that active case finding was more effective at reducing the prevalence of TB disease than current passive finding measures. Reducing TB burden may rely on actively screening endemic communities to identify infectious individuals and initiate early treatment, especially as evidence suggests that individuals with active TB disease may remain asymptomatic and infectious for a considerable period of time. 2 , 3
To actively screen communities for TB, the diagnostic test should be rapid, relatively cheap and ideally non‐sputum‐based. Existing tools for the identification of TB rely on the analysis of sputum for Mycobacterium tuberculosis by either microscopy, culture or PCR. Expectorating sputum increases the risk of transmission, and good quality sputum can be challenging to obtain from children and the elderly. The recent study by Marks et al. 2 reported that only half the study participants could produce a quality sputum sample for Xpert analysis. Nor is sputum useful for identifying extrapulmonary TB, which accounts for 15–30% of TB cases. 4 , 5 Alternative biological samples for TB diagnosis, such as urine, saliva and blood, are of increasing research interest for the development of new tests for TB diagnosis.
Efforts to identify M. tuberculosis or mycobacterial components from non‐sputum biological samples have been largely unsuccessful to date, owing to a lack of sensitivity and specificity. 6 , 7 Other measurable changes in the host response may aid in developing a new non‐sputum test for screening for active TB. Host biomarkers are an attractive option for new TB diagnostics. Many candidates are currently being explored, with whole‐blood or plasma mRNA, 8 , 9 proteins 10 and microRNA (miRNA) 11 being considered. This study builds on our previous study 12 that demonstrated the biomarker potential of a five miRNA signature to identify active TB disease. The utility of host protein biomarkers in plasma for the identification of active TB and to monitor response to therapy in a cohort of TB patients, alone or in combination with the previously identified miRNA biosignature was examined. Host proteins are readily measured in the circulation and are relatively stable. 13 Cytokines, chemokines and growth factors are known to change systemically during TB infection and provide promising candidates. 10 , 14 , 15 IFNγ is crucial for TB disease control, yet biomarker studies have indicated that this cytokine alone does not have the diagnostic accuracy for a point‐of‐care test for active TB. 16 , 17 By contrast, the CXCL10 chemokine, interferon‐inducible protein‐10 (IP‐10), has emerged as a leading candidate as a biomarker for active TB disease, although additional studies with larger sample sizes in different regions are required to confirm these findings. 16 , 18 , 19 , 20 , 21 , 22 To investigate this, we selected the following protein panel based on the diagnostic potential of the components in previous studies: IP‐10, 16 , 18 , 19 , 20 , 21 , 22 CCL2 (monocyte chemoattractant protein‐1, MCP‐1), 23 CCL5 (RANTES), 24 CCL11 (Eotaxin), 25 , 26 TNF, 27 , 28 soluble TNF receptor 1 (sTNFR1), 29 IL‐6, 17 , 24 , 27 IL‐10 17 , 27 and vascular endothelial growth factor (VEGF). 22 , 28
The potential of these nine host proteins as a biosignature to distinguish active TB patients from controls was investigated in a cohort of 100 pulmonary TB patients. The biomarker potential of these proteins was then compared to that of the miRNA signature previously shown to have good sensitivity and specificity at distinguishing active TB from healthy controls. All individual samples were from the Ningxia Hui Autonomous Region (NHAR), China, where the TB prevalence was estimated to be 61 cases per 100 000 between 2005 and 2008, although rates are predicted to be higher. 30 The prognostic potential of these proteins as indicators of the treatment response was also examined and the biomarker potential of a novel combined miRNA and protein biosignature for active TB described.
Results
Protein expression in newly diagnosed TB patients
Samples were obtained from consenting TB patients and control subjects recruited from NHAR. All individuals were over the age of 18 and had no history of previous TB disease diagnosis or treatment, prior to this presentation. Patients and controls were of either Han or Hui ethnicity and included both males and females (Supplementary table 1). The expression of nine proteins in the plasma of 100 newly diagnosed TB patients (treatment naïve) and 100 controls, matched for age and sex, was assessed. Six of these proteins (IP‐10, MCP‐1, sTNFR1, RANTES, VEGF and IL‐6) were significantly increased in the plasma of TB patients than controls, while TNF levels were significantly decreased (Figure 1). Data from the TB patients and control subjects were then analysed stratified on the basis of gender and ethnicity (Han or Hui). No significant differences in protein expression were observed between males and females or TB patients from different ethnic backgrounds (Supplementary figures 1 and 2).
Figure 1.
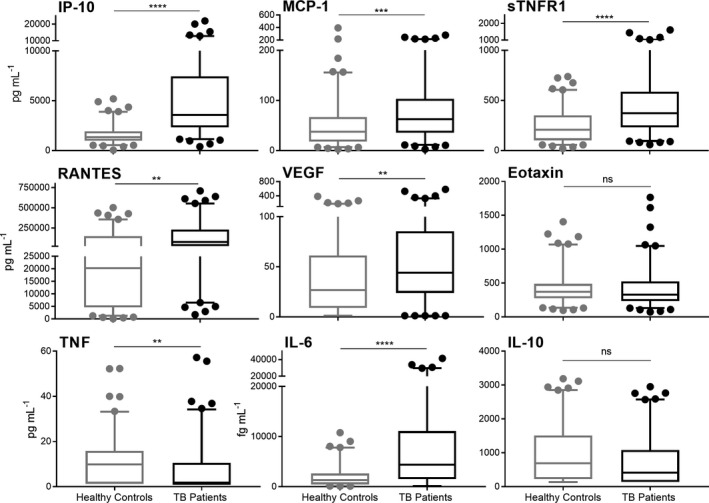
Expression of nine plasma proteins in 100 TB patients at diagnosis (treatment naïve) compared to age‐ and sex‐matched healthy controls. Data are represented by boxplots with median and interquartile range with bars from the 5–95 percentile. Samples were statistically analysed by Welch’s t‐test (ns = no significance, **P‐value < 0.01, ***P‐value < 0.001, ****P‐value < 0.0001).
To determine the sensitivity and specificity of these proteins to distinguish TB patients from healthy controls at diagnosis, receiver operator characteristic (ROC) curves were generated for single proteins (Figure 2). IP‐10 was the protein with the highest area under the curve (AUC) of 0.874, in distinguishing TB patients from controls with a sensitivity and specificity of 75% and 87% respectively. To examine whether the accuracy of individual proteins could be improved, ROC curve analysis on combinations of several proteins was assessed. Together, the nine proteins could distinguish TB patients from controls with an AUC = 0.908 and sensitivity of 80% and specificity of 88%. The effect of varying the number of proteins in the panel was examined by binary logistic regression. This indicated that a panel of three proteins (IP‐10, TNF and Eotaxin) was sufficient to maintain the same level of sensitivity and specificity while only marginally decreasing the AUC to 0.903 (Figure 3 and Table 1).
Figure 2.
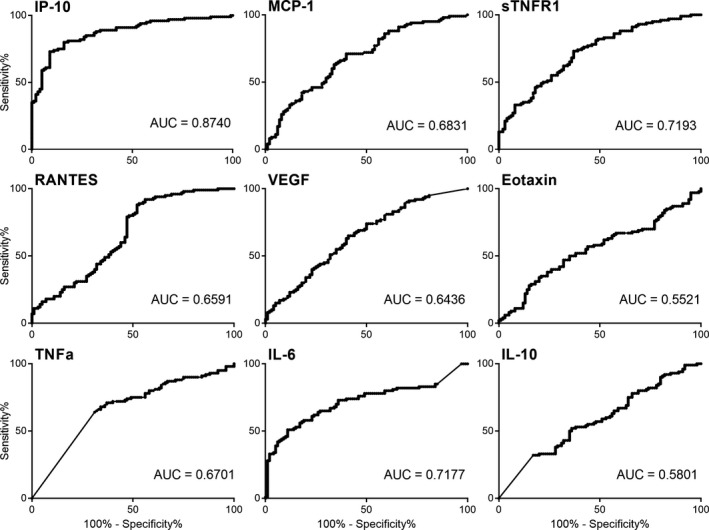
ROC curves and AUC values for the nine individual proteins to distinguish TB patients at time of diagnosis from controls. IP‐10 plasma levels generated the best diagnostic accuracy from the nine proteins with an AUC = 0.874. AUC, area under the curve; ROC, receiver operator characteristic.
Figure 3.
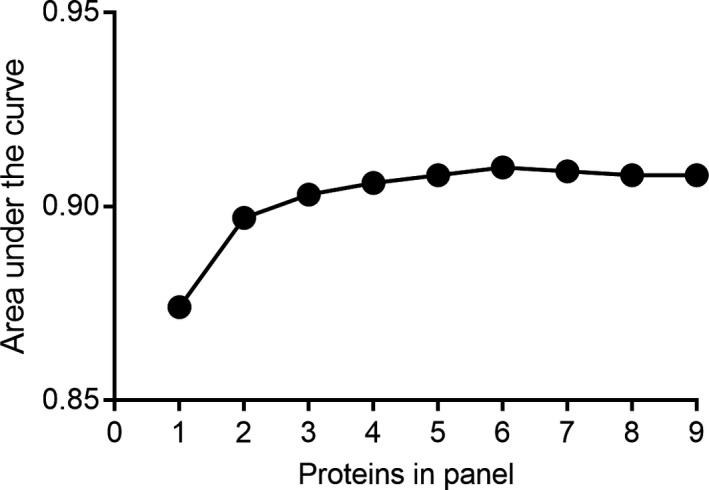
The area under the curve for increasing numbers of plasma proteins in the biosignature for distinguishing pulmonary TB patients at the time of diagnosis from control subjects, as determined binary logistic regression.
Table 1.
Sensitivity, specificity, negative predictive value and positive predictive value for increasing numbers of proteins in a biosignature panel, to differentiate TB patients from controls
| Number of proteins in panel | Proteins added to panel | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|
| 1 | IP‐10 | 75.00 | 87.00 | 85.23 | 77.68 |
| 2 | TNF | 79.00 | 85.00 | 84.04 | 80.19 |
| 3 | Eotaxin | 80.00 | 88.00 | 86.96 | 81.48 |
| 4 | RANTES | 79.00 | 87.00 | 85.87 | 80.56 |
| 5 | IL‐6 | 79.00 | 86.00 | 84.95 | 80.37 |
| 6 | sTNFR1 | 82.00 | 87.00 | 86.32 | 82.86 |
| 7 | MCP‐1 | 80.00 | 87.00 | 86.02 | 81.31 |
| 8 | IL‐10 | 80.00 | 88.00 | 86.96 | 81.48 |
| 9 | VEGF | 80.00 | 88.00 | 86.96 | 81.48 |
Changes in plasma protein expression during treatment
The capacity of the expression levels of these nine proteins to identify response to therapy of TB patients during their treatment course was then assessed. Samples taken at 1, 2 and 6 months during standard therapy were measured. Interestingly, only IP‐10 and IL‐6 levels significantly decreased within the first month of therapy and continued to trend towards levels in healthy control subjects during treatment (Figure 4). All of the proteins that were significantly elevated at diagnosis remained increased compared to the levels in healthy controls at the end of treatment, except for IL‐6 (Figure 4).
Figure 4.
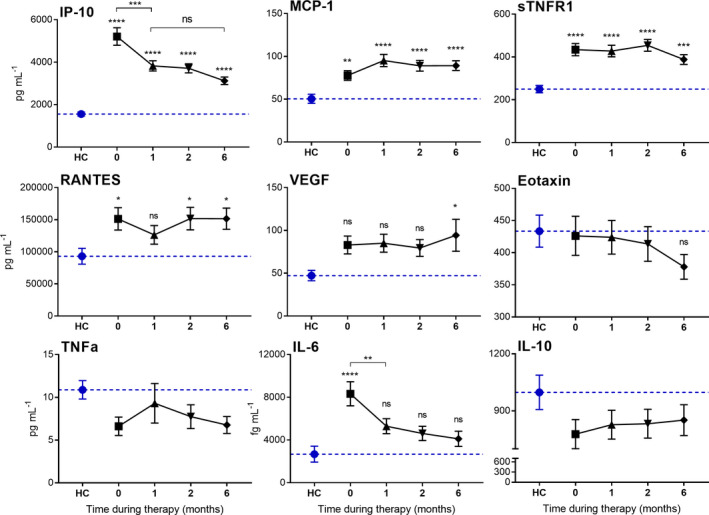
Expression of nine plasma proteins in TB patients during 6 months of standard TB treatment and in healthy control subjects (HC). Data are mean ± SEM, n = 100 per group. The dotted line is representative of the mean protein level for the HC cohort. Statistical differences in levels compared to healthy controls were assessed by one‐way ANOVA with Tukey’s multiple comparisons test (ns = no significance, *P‐value < 0.05, **P‐value < 0.01, ***P‐value < 0.001, ****P‐value < 0.0001).
Plasma protein levels at diagnosis (time 0) were stratified based on whether the patient successfully completed recommended treatment. Treatment failure was determined by a positive sputum culture at the end of 6‐month standard therapy and/or progressive radiological changes. Eleven TB patients out of the 100 were considered to have failed therapy. As shown in Figure 5, these 11 TB patients had significantly elevated IL‐6 and RANTES plasma levels at diagnosis when compared to the TB patients that successfully completed therapy.
Figure 5.
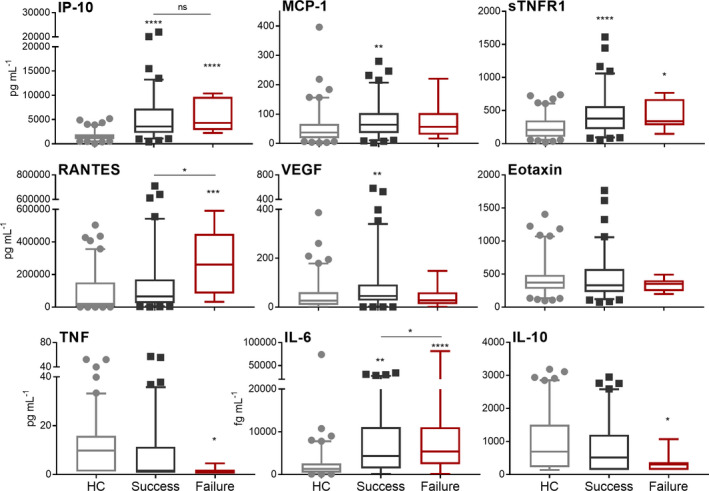
Plasma protein levels in TB patients at time of diagnosis (treatment naïve) who successfully completed standard therapy (n = 89, black) or who failed therapy (n = 11, red) compared to levels in controls (n = 100, grey). Statistical differences were assessed by one‐way ANOVA with Tukey’s multiple comparisons test. Significant differences displayed are relative to the HC group unless otherwise indicated (ns = no significance, *P‐value < 0.05, **P‐value < 0.01, ***P‐value < 0.001, ****P‐value < 0.0001).
Protein and miRNA
In addition to the nine proteins measured in this study, our previous study 12 measured the biomarker potential of a panel of 10 miRNA in these same plasma samples by qPCR. The expression levels for all nine proteins and 10 miRNA were collated. Analysis of all 19 analytes together demonstrated an AUC = 1.000 with sensitivity of 100% and specificity of 100% for distinguishing TB patients from control subjects at diagnosis (Supplementary table 2). As shown in Figure 6, reducing the number of miRNA and proteins in this combined panel to just five analytes (IP‐10, miR‐29a, miR‐146a, miR‐99b and miR‐221) still gave an AUC of 0.995 with a sensitivity of 96% and specificity of 97%. IP‐10 was the most accurate predictor of TB disease from all 19 plasma miRNA and proteins.
Figure 6.
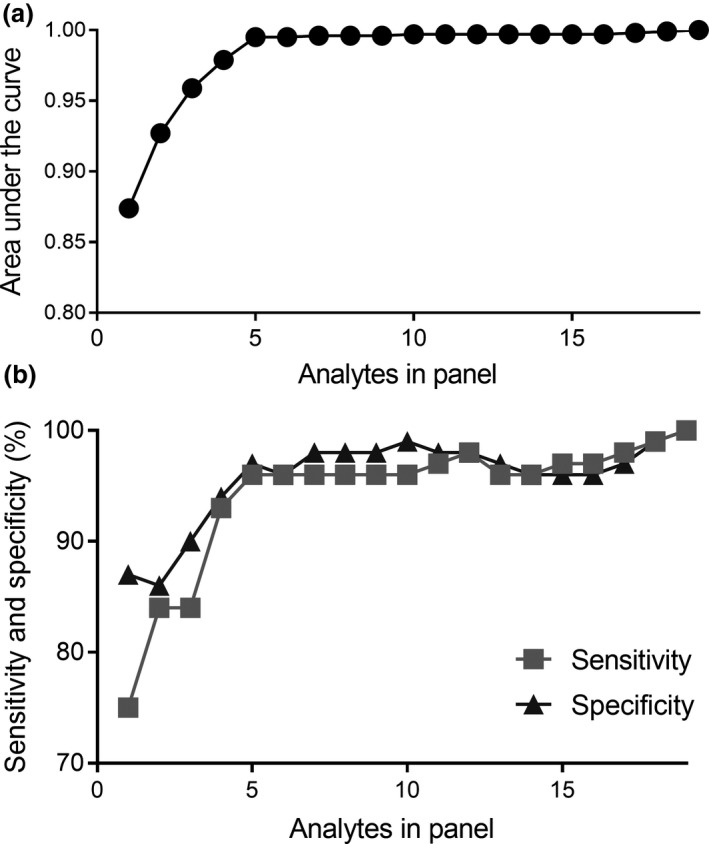
The area under the curve (a), sensitivity and specificity (b) for varying the number of plasma analytes in a biomarker signature to distinguish active TB patients from control subjects. The 19 analytes consisted of nine proteins measured in this study and 10 miRNA reported in a previous study. 12 The best single biomarker candidate was IP‐10.
Discussion
Decreasing the TB burden globally requires new diagnostic tools. A simple, rapid test that does not rely on sputum would greatly benefit this goal. Our study identified a 3‐protein biomarker panel that distinguishes individuals with active TB disease from controls with an AUC = 0.903 and a high sensitivity and specificity of 80% and 88%, respectively. Interestingly, most of this diagnostic accuracy was because of the single plasma protein IP‐10.
IP‐10 was significantly up‐regulated in the plasma of Chinese TB patients compared to controls. IP‐10 has been identified in previous studies as a biomarker candidate for diagnosing active TB disease. 21 , 31 , 32 , 33 , 34 Studies in both low and high TB endemic regions including China, Uganda, Norway and Denmark have demonstrated that IP‐10 in blood, plasma or serum was significantly up‐regulated in TB patients in comparison with control cohorts. 31 , 32 , 33 , 34 Alone, however, IP‐10 does not have the sensitivity required for a non‐sputum biomarker test for active TB disease, which according to the WHO requirements is ≥ 90%. 35 ROC curve analysis of IP‐10 in previous biomarker studies ranged from 0.801 to 0.950 and sensitivity ranged from 72.4 to 83.3%, 31 , 32 , 34 which is in accordance with our study.
Changes in plasma IP‐10 levels during infection are not unique to TB disease. IP‐10 is a chemokine associated with inflammation. An increase in IP‐10 levels has shown biomarker potential in a number of TB biomarker studies. 19 , 21 , 36 Increased IP‐10 has also been associated with a number of other infections in adults with respiratory viruses such as influenza, 37 and with severe malaria 38 and in cystic fibrosis patients during pulmonary exacerbations. 39 Binary logistic regression showed that increased IP‐10, together with TNF and Eotaxin levels, was able to identify TB patients from control subjects with a high sensitivity and specificity, despite Eotaxin levels not differing significantly different between the two groups. This may indicate a biological link between the levels of these three proteins during TB disease progression.
Practically, however, increases in IP‐10 and IL‐6 plasma levels, which were able to identify TB patients with an AUC = 0.875, a sensitivity of 75% and specificity of 87%, represent changes that may be easier to measure as indicators of inflammation and disease. These data suggest a role for IP‐10 as a triage tool whereby high levels are indicative of active infection and the requirement for further testing. Utilising a plasma IP‐10 cut‐off level of 1000 pg mL−1, whereby any individual with higher levels would be deemed positive, identifies 97% of all TB patients in this cohort. This equates to a false‐negative rate of only 3%, although a high false‐positive rate of 72%. Adding IL‐6 to this test, with a cut‐off level of 3000 fg mL−1, lowers the false‐positive rate to 12%. If applied as a triage tool, high expression of both IP‐10 and IL‐6 is strongly indicative of TB disease and could be used to trigger further microbiological testing.
One way to improve the sensitivity of biomarkers may be the addition of multiple analytes 25 , 26 , 40 or to expand the types of biomarkers within the panel. Our previous study showed that a panel of five miRNA could identify active TB patients with a sensitivity and specificity of 94% and 88%, respectively. 12 Additional studies have also identified the potential of plasma miRNAs as biomarkers for active TB disease. 11 Therefore, the biomarker potential of a combined miRNA and protein panel was assessed. Analysing these data using the combination of 19 miRNA and protein analytes greatly improved diagnostic accuracy with an AUC of 1.00, sensitivity and specificity of 100%. A panel of 19 markers is unrealistic for development as a biomarker test. Reducing this panel to five analytes (IP‐10, miR‐29a, miR‐146a, miR‐99b and miR‐221) maintained good accuracy with an AUC = 0.995, sensitivity of 96% and specificity of 97%. These data indicate that a novel combination of miRNA and protein biomarkers was able to improve the accuracy of either proteins or miRNA biomarker candidates alone. It is increasingly clear that multiple biomarkers will be required to reach the necessary high sensitivity and specificity outlined by the WHO, 41 and this novel combination may be a promising avenue for active TB biomarker development.
Using proteins as biomarkers for a non‐sputum test is attractive as proteins are relatively stable in the circulation and easy to measure with limited tools. While miRNA are also considered relatively stable in the circulation, they are more difficult to measure and require PCR technology. Requiring the simultaneous measurement of both protein and miRNA is a drawback for developing this biomarker test. As technology advances, new techniques capable of measuring miRNA in bead‐based assays are being developed 42 and there is the potential to combine the measurement of both miRNA and protein on a similar platform.
One requirement for advancing a biomarker test for active TB is developing a universal test with diagnostic accuracy across multiple ethnicities and geographical locations. In this study, the biomarker panel was equally effective in identifying TB disease in both males and females, and in the Han and Hui subjects (Supplementary figures 1 and 2), further studies in larger diverse populations at multiple locations will be required to determine the effectiveness of this panel as a universal diagnostic tool. There is a growing body of literature assessing host proteins as biomarkers of active TB disease. 43 The lack of validation of candidates between populations, and differences in sample choice, preparation and processing are all inhibiting progress in diagnostic development. There is a need to assess the potential of these biomarker candidates in different ethnic and geographical populations to identify both baseline and levels associated with disease. A recent review acknowledged that one shortcoming to advancing this field is the availability of data. 43 It proposed a TB biomarker database as an efficient and necessary tool to summarise the current findings and match studies with similar biomarker candidates to improve validation and the development of a test for active TB. Developing systems to share biomarker data more efficiently and confirm results in multiple populations is essential to progress biomarker development.
Another consideration for TB biomarker development is the expression profile of individuals with LTBI. It is estimated that almost a quarter of the global population have LTBI. 44 Most will not develop active TB disease and do not require treatment. 45 However, studies have shown that in the 6 months before reactivation there are measurable changes in the whole‐blood mRNA profiles of reactivating LTBI‐infected individuals. 46 Reactivation of LTBI infection may also modify the miRNA and protein expression profiles and this may aid early diagnosis. Future biomarker studies could include analysis of reactivation within a LTBI cohort.
One strength of this study was that TB patients were monitored throughout their treatment, providing samples during therapy. This allowed us to identify how the levels of these inflammatory proteins change with treatment and recovery. There is growing interest in biomarkers to not only identify TB disease but also to monitor the response to therapy and so rapidly identify patients who are not responding to treatment. This failure to respond may be caused by drug resistance, poor compliance or poor drug absorption. Currently, TB cure is defined as being sputum culture negative at the end of intensive phase (2 months) and at treatment completion. Interestingly, of the 6 proteins significantly elevated at diagnosis in TB patients, only two, IP‐10 and IL‐6, declined significantly over the 6‐month therapy, and only IL‐6 returned to baseline levels. In another study, IP‐10 was similarly shown to be reduced by therapy. 22 This study also found that VEGF fell significantly during treatment, but this finding was not reproduced by our study. Further initial levels of IL‐6 and RANTES were significantly higher in the plasma of TB patients who went on to fail therapy than in patients who successfully completed treatment. These proteins may have potential as early indicators of treatment outcome. However, the group of TB patients that failed therapy was small with only 11 individuals, and further research with larger cohorts is required to validate these findings. Four proteins (MCP‐1, RANTES, sTNFR1 and VEGF) that were elevated pre‐treatment remained significantly elevated upon the conclusion of treatment, indicating that the extensive inflammation and lung remodelling that occurs during TB disease is continuing even at the completion of successful treatment. 47 This is consistent with the continuing inflammation identified by PET/CT scanning of pulmonary TB patients at the completion of therapy. 48
It is a strength of this study to have sampled plasma from the TB patients four times throughout treatment, and a limitation that control subjects were only sampled once, so we are unable to assess variation in the levels of these plasma proteins in controls over time. Further research is needed to determine the individual variation in baseline plasma protein levels and how this may impact the utility of these proteins as indicators of treatment response. There is also the need to determine the biomarker signature of those with other respiratory diseases to determine whether these proteins, either as a triage tool or as a point‐of‐care test, could distinguish active TB from other diseases with similar symptoms. The method used to diagnose individuals with active TB disease followed national guidelines. 33% of these individuals had microbiological confirmed sputum smear of M. tuberculosis. At the time of sample collection, GeneXpert analysis was not available in the NHAR. Future work should address these weaknesses to strengthen the research in this area.
Conclusion
Plasma proteins are a viable option as potential biomarkers of active TB disease with IP‐10 the leading candidate with an AUC = 0.874, although a biomarker panel or signature of multiple analytes is more likely required to achieve a better diagnostic accuracy. Plasma IP‐10 may have an important role as a triage tool, identifying patients that require further investigation to exclude TB. This study demonstrates that a novel combination of miRNA and protein biomarkers performed better than either proteins or miRNA biomarker panels alone. Further validation of these biomarker candidates in other geographical or ethnic populations is required to advance TB biomarker development.
Methods
Study population
All participants were recruited through the TB Control Program in the NHAR, China, 12 and TB patients were diagnosed based on national guidelines. 30 National guidelines for TB detection involve passive case finding, whereby individuals with symptoms seek care. A chest X‐ray was performed for every patient and control subject recruited. These were read by two independent medical specialists, and all patients had radiological findings in keeping with active pulmonary TB. Sputum analysis including smear microscopy and culture was performed where samples were available. Thirty‐three patients had culture‐positive sputum, in line with data from NHAR, where culture‐positive TB was identified in approximately one‐third of new patients. The diagnosis of TB was made on the basis of clinical suspicion, radiological findings and sputum smear and culture. In sputum culture‐negative cases, clinical response to therapy coupled with radiological improvement was used to confirm the initial diagnosis of pulmonary TB. Ethical approval for this study was granted from the University of Sydney Ethics Review Committee (protocol no. 2012/1076) and the Ningxia Medical University Human Ethics Committee (approval date 6/6/2013). All subjects gave written informed consent before their enrolment in the study.
Plasma samples were collected from 100 TB patients at diagnosis (treatment naïve) and 100 healthy, age‐ and gender‐matched controls. The characteristics of participants are summarised in Supplementary table 1. The TB patients were initiated on standard TB treatment according to national guidelines that follow WHO DOTS guidelines for 6 months. Additional blood samples were collected from patients at 1, 2 and 6 months during treatment.
Sample collection and preparation
Ten millilitres of venous blood was collected into an EDTA tube. 12 Blood was processed within 2 h of collection. Blood was separated by centrifugation at 800 g for 10 min. Plasma was removed and stored at –80°C. Samples were examined for haemolysis using a haemolysis chart and samples with > 100 mg dL−1 of haemoglobin were discarded.
Protein expression
Protein expression was detected in plasma samples by cytometric bead array (CBA) (BD Biosciences, Australia). IL‐6 and IL‐10 were assessed with an enhanced sensitivity CBA, as per the manufacturer’s protocol, with a lower detection limit of 274 fg mL−1. IP‐10, MCP‐1, RANTES, TNF, sTNFR1, Eotaxin and VEGF levels were measured with the CBA array kit, with a detection limit of 2.5 pg mL−1, as per the manufacturer’s instructions. Plasma samples were diluted 1:4 as recommended for protein quantification. RANTES expression was higher than the array’s upper limit of detection of 2500 pg mL−1, and therefore, the plasma samples were diluted 1:200 for RANTES quantification. Any samples that fell below the array’s lower limit of detection were allocated a value equal to the half limit of detection. Samples were acquired by flow cytometer, either a BD Fortessa X20 (University of Technology Sydney) or BD Canto‐II (Centenary Institute).
Statistical analysis
Flow cytometry files were analysed by FCAP Array software (BD Biosciences, Australia). Statistical analysis was in GraphPad Prism v7 by Welch's t‐test or one‐way ANOVA with Tukey’s multiple comparison test.
Receiver operating characteristic (ROC) curves for single proteins were analysed in GraphPad Prism v7. ROC curves show the relationship between clinical sensitivity and specificity. They are generated by plotting the sensitivity and specificity of data based on a series of cut‐off values. Sensitivity refers to the rate of true positives (TB patients) correctly identified. Specificity is the rate of true negatives (control subjects) correctly identified. For the resulting ROC curve generated, an AUC value is calculated. An AUC = 1.00 demonstrates that the test is 100% accurate and every positive or negative sample is correctly identified. The positive predictive value (PPV) and negative predictive value (NPV) of each biomarker were examined. PPV refers to the probability that a positive test result is a true TB patient whereas NPV refers to the probability that a negative test result does not have TB.
Receiver operator characteristic curves for both protein panels and combined miRNA and protein panels were generated in IBM SPSS Statistics 25 software following binary logistic regression calculations.
The protein data set generated in this study was then combined with our miRNA data set (10 miRNA – confirmatory study) previously published by Barry et al. 2018 which describes the sample collection and a miRNA profiling biomarker study on these samples. 12 miRNA biomarker candidates were miR‐21‐5p; −99b‐5p; −29a‐5p; −223‐5p; −221‐3p; −146a‐5p; −26a‐5p; −28‐5p; −133a; and −652‐3p.
Conflict of interest
The authors report no conflicts of interest with regard to this publication.
Author contribution
Jessica L Pedersen: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing‐original draft. Simone E Barry: Data curation; Investigation; Writing‐review & editing. Nilesh J Bokil: Conceptualization; Project administration; Supervision; Writing‐review & editing. Magda Ellis: Data curation; Writing‐review & editing. YuRong Yang: Investigation; Resources; Writing‐review & editing. Guangyu Guan: Investigation; Resources; Writing‐review & editing. Xiaolin Wang: Investigation; Resources; Writing‐review & editing. Alen Faiz: Software; Writing‐review & editing. Warwick Britton: Conceptualization; Project administration; Writing‐review & editing. Bernadette M Saunders: Conceptualization; Funding acquisition; Project administration; Supervision; Writing‐review & editing.
Supporting information
Acknowledgments
JP, NB, BS proposed the experimental design. JP executed the experiments. BS, SB, YY, GG, XW and WB undertook the collection of all plasma samples. SB, BS, ME and WB designed the miRNA profiling experiments, 12 of which the miRNA confirmatory dataset was used in this study. AF helped with data interpretation and analysis. JP and BS composed the manuscript. All authors reviewed and edited the manuscript. This work was supported by the National Health and Medical Research Council – Centres for Research Excellence in Tuberculosis Control [APP1043225, APP1153493]. JP was supported by an Australian Government Research Training Program Scholarship.
References
- 1. WHO . Global Tuberculosis Report 2020. World Health Organization, 2020. [Google Scholar]
- 2. Marks GB, Nguyen NV, Nguyen PTB et al. Community‐wide Screening for Tuberculosis in a High‐Prevalence Setting. N Engl J Med 2019; 381: 1347–1357. [DOI] [PubMed] [Google Scholar]
- 3. Drain PK, Bajema KL, Dowdy D et al. Incipient and subclinical tuberculosis: a clinical review of early stages and progression of infection. Clin Microbiol Rev 2018; 31: e00021‐00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sandgren A, Hollo V, van der Werf MJ . Extrapulmonary tuberculosis in the European Union and European Economic Area, 2002 to 2011. Euro Surveill 2013; 18: e24031. [PubMed] [Google Scholar]
- 5. Gaifer Z. Epidemiology of extrapulmonary and disseminated tuberculosis in a tertiary care center in Oman. Int J Mycobacteriol 2017; 6: 162–166. [DOI] [PubMed] [Google Scholar]
- 6. Hamasur B, Bruchfeld J, Haile M et al. Rapid diagnosis of tuberculosis by detection of mycobacterial lipoarabinomannan in urine. J Microbiol Methods 2001; 45: 41–52. [DOI] [PubMed] [Google Scholar]
- 7. Al‐Zamel FA. Detection and diagnosis of Mycobacterium tuberculosis . Expert Rev Anti Infect Ther 2009; 7: 1099–1108. [DOI] [PubMed] [Google Scholar]
- 8. Warsinske H, Vashisht R, Khatri P. Host‐response‐based gene signatures for tuberculosis diagnosis: a systematic comparison of 16 signatures. PLoS Med 2019; 16: e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ho J, Bokil NJ, Nguyen PTB et al. A transcriptional blood signature distinguishes early tuberculosis disease from latent tuberculosis infection and uninfected individuals in a Vietnamese cohort. J Infect 2020; 81: 72–80. [DOI] [PubMed] [Google Scholar]
- 10. Won EJ, Choi JH, Cho YN et al. Biomarkers for discrimination between latent tuberculosis infection and active tuberculosis disease. J Infect 2017; 74: 281–293. [DOI] [PubMed] [Google Scholar]
- 11. Pedersen JL, Bokil NJ, Saunders BM. Developing new TB biomarkers, are miRNA the answer? Tuberculosis 2019; 118: 101860. [DOI] [PubMed] [Google Scholar]
- 12. Barry SE, Ellis M, Yang Y et al. Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti‐mycobacterial therapy. J Infect 2018; 77: 341–348. [DOI] [PubMed] [Google Scholar]
- 13. Graham C, Chooniedass R, Stefura WP et al. Stability of pro‐ and anti‐inflammatory immune biomarkers for human cohort studies. J Transl Med 2017; 15: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo J, Zhang M, Yan B et al. Diagnostic performance of plasma cytokine biosignature combination and MCP‐1 as individual biomarkers for differentiating stages Mycobacterium tuberculosis infection. J Infect 2019; 78: 281–291. [DOI] [PubMed] [Google Scholar]
- 15. La Manna MP, Orlando V, Li Donni P et al. Identification of plasma biomarkers for discrimination between tuberculosis infection/disease and pulmonary non tuberculosis disease. PLoS One 2018; 13: e0192664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei M, Wu ZY, Lin JH et al. Regulation network of serum cytokines induced by tuberculosis‐specific antigens reveals biomarkers for tuberculosis diagnosis. Genet Mol Res 2015; 14: 17182–17192. [DOI] [PubMed] [Google Scholar]
- 17. Kisuya J, Chemtai A, Raballah E, Keter A, Ouma C. The diagnostic accuracy of Th1 (IFN‐γ, TNF‐α, and IL‐2) and Th2 (IL‐4, IL‐6 and IL‐10) cytokines response in AFB microscopy smear negative PTB‐ HIV co‐infected patients. Sci Rep 2019; 9: e2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wergeland I, Pullar N, Assmus J et al. IP‐10 differentiates between active and latent tuberculosis irrespective of HIV status and declines during therapy. J Infect 2015; 70: 381–391. [DOI] [PubMed] [Google Scholar]
- 19. Petrone L, Cannas A, Vanini V et al. Blood and urine inducible protein 10 as potential markers of disease activity. Int J Tuberc Lung Dis 2016; 20: 1554–1561. [DOI] [PubMed] [Google Scholar]
- 20. Mamishi S, Mahmoudi S, Banar M, Hosseinpour Sadeghi R, Marjani M, Pourakbari B. Diagnostic accuracy of interferon (IFN)‐γ inducible protein 10 (IP‐10) as a biomarker for the discrimination of active and latent tuberculosis. Mol Biol Rep 2019; 46: 6263–6269. [DOI] [PubMed] [Google Scholar]
- 21. Jacobs R, Malherbe S, Loxton AG et al. Identification of novel host biomarkers in plasma as candidates for the immunodiagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. Oncotarget 2016; 7: 57581–57592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Riou C, Perez Peixoto B, Roberts L et al. Effect of standard tuberculosis treatment on plasma cytokine levels in patients with active pulmonary tuberculosis. PLoS One 2012; 7: e36886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiong W, Dong H, Wang J et al. Analysis of plasma cytokine and chemokine profiles in patients with and without tuberculosis by liquid array‐based multiplexed immunoassays. PLoS One 2016; 11: e0148885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Y, Yang X, Zhang X et al. IP‐10 and RANTES as biomarkers for pulmonary tuberculosis diagnosis and monitoring. Tuberculosis 2018; 111: 45–53. [DOI] [PubMed] [Google Scholar]
- 25. Chen T, Li Z, Yu L et al. Profiling the human immune response to Mycobacterium tuberculosis by human cytokine array. Tuberculosis 2016; 97: 108–117. [DOI] [PubMed] [Google Scholar]
- 26. Choi R, Kim K, Kim MJ et al. Serum inflammatory profiles in pulmonary tuberculosis and their association with treatment response. J Proteomics 2016; 149: 23–30. [DOI] [PubMed] [Google Scholar]
- 27. Bai R, Tao L, Li B et al. Using cytometric bead arrays to detect cytokines in the serum of patients with different types of pulmonary tuberculosis. Int J Immunopathol Pharmacol 2019; 33: 2058738419845176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hur YG, Kang YA, Jang SH et al. Adjunctive biomarkers for improving diagnosis of tuberculosis and monitoring therapeutic effects. J Infect 2015; 70: 346–355. [DOI] [PubMed] [Google Scholar]
- 29. Kawaguchi H, Ina Y, Ito S et al. Serum levels of soluble tumor necrosis factor (TNF) receptors in patients with pulmonary tuberculosis. Kekkaku 1996; 71: 259–265. [PubMed] [Google Scholar]
- 30. Yang YR, McManus DP, Gray DJ et al. Evaluation of the tuberculosis programme in Ningxia Hui Autonomous region, the People's Republic of China: a retrospective case study. BMC Public Health 2012; 12: 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang Y, Wang Y, Li H et al. Evaluation of a whole‐blood chemiluminescent immunoassay of IFN‐γ, IP‐10, and MCP‐1 for diagnosis of active pulmonary tuberculosis and tuberculous pleurisy patients. APMIS 2016; 10: 856–864. [DOI] [PubMed] [Google Scholar]
- 32. Biraro IA, Kimuda S, Egesa M et al. The Use of interferon gamma inducible protein 10 as a potential biomarker in the diagnosis of latent tuberculosis infection in Uganda. PLoS One 2016; 11: e0146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tonby K, Ruhwald M, Kvale D, Dyrhol‐Riise AM. IP‐10 measured by dry plasma spots as biomarker for therapy responses in Mycobacterium Tuberculosis infection. Sci Rep 2015; 5: e9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruhwald M, Bodmer T, Maier C et al. Evaluating the potential of IP‐10 and MCP‐2 as biomarkers for the diagnosis of tuberculosis. Eur Respir J 2008; 32: 1607–1615. [DOI] [PubMed] [Google Scholar]
- 35. WHO . High priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting, 28‐29 April 2014, Geneva, Switzerland. World Health Organization; 2014. [Google Scholar]
- 36. Hong JY, Jung GS, Kim H et al. Efficacy of inducible protein 10 as a biomarker for the diagnosis of tuberculosis. Int J Infect Dis 2012; 16: e855–e859. [DOI] [PubMed] [Google Scholar]
- 37. Hayney MS, Henriquez KM, Barnet JH et al. Serum IFN‐γ‐induced protein 10 (IP‐10) as a biomarker for severity of acute respiratory infection in healthy adults. J Clin Virol 2017; 90: 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jain V, Armah HB, Tongren JE et al. Plasma IP‐10, apoptotic and angiogenic factors associated with fatal cerebral malaria in India. Malar J 2008; 7: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Solomon GM, Frederick C, Zhang S et al. IP‐10 is a potential biomarker of cystic fibrosis acute pulmonary exacerbations. PLoS One 2013; 8: e72398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chegou NN, Sutherland JS, Malherbe S et al. Diagnostic performance of a seven‐marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax 2016; 71: 785–794. [DOI] [PubMed] [Google Scholar]
- 41. Denkinger CM, Schumacher SG, Gilpin C et al. Guidance for the evaluation of tuberculosis diagnostics that meet the World Health Organization (WHO) target product profiles: an introduction to WHO process and study design principles. J Infect Dis 2019; 220: S91–S98. [DOI] [PubMed] [Google Scholar]
- 42. Biscontin A, Casara S, Cagnin S et al. New miRNA labeling method for bead‐based quantification. BMC Mol Biol 2010; 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacLean E, Broger T, Yerlikaya S, Fernandez‐Carballo BL, Pai M, Denkinger CM. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol 2019; 4: 748–758. [DOI] [PubMed] [Google Scholar]
- 44. Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re‐estimation using mathematical modelling. PLoS Med 2016; 13: e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marks GB, Bai J, Simpson SE, Sullivan EA, Stewart GJ. Incidence of tuberculosis among a cohort of tuberculin‐positive refugees in Australia: reappraising the estimates of risk. Am J Respir Crit Care Med 2000; 162: 1851–1854. [DOI] [PubMed] [Google Scholar]
- 46. Zak DE, Penn‐Nicholson A, Scriba TJ et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 2016; 387: 2312–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dorhoi A, Kaufmann SH. Pathology and immune reactivity: understanding multidimensionality in pulmonary tuberculosis. Semin Immunopathol 2016; 38: 153–166. [DOI] [PubMed] [Google Scholar]
- 48. Malherbe ST, Shenai S, Ronacher K et al. Persisting positron emission tomography lesion activity and Mycobacterium tuberculosis mRNA after tuberculosis cure. Nat Med 2016; 22: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


