Fig. 3.
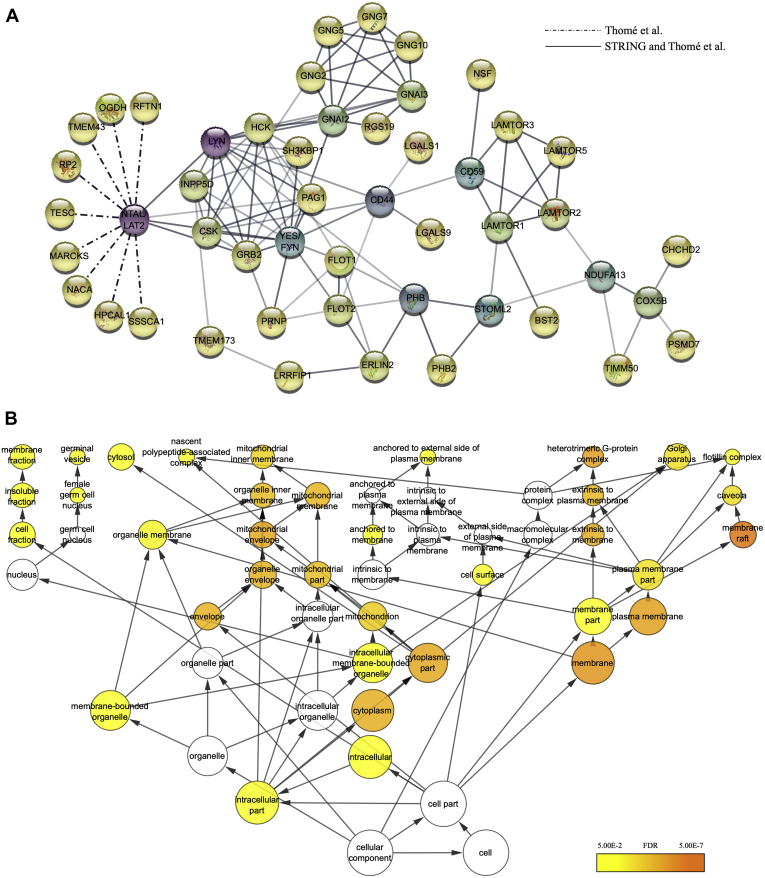
Bioinformatics analysis of NTAL protein interactors identified by IP–MS/MS (IP/MS).A, STRING (https://string-db.org/) and Cytoscape software were used for the functional analysis of protein–protein interactions. Proteins in purple, blue, and green spheres represent the first shelf of protein interactions, whereas proteins in yellow spheres represent the second shelf of interactors. Dashed lines indicates protein interactions only described by the current study, whereas continuous lines represent protein interactions that were previously described and also observed in the current study. B, GO analysis of NTAL interactors identified by MS analysis in NB4 and U937 cells was performed using BiNGO analysis of Cytoscape software. Yellow circles represent the GO terms statistically significant. GO terms for cellular component (CC) with a statistical significance of FDR q value <0.05 were considered for further conclusions. BiNGO, Biological Networks Gene Ontology; FDR, false discovery rate; GO, Gene Ontology; IP, immunoprecipitation; NTAL, non–T cell activation linker.