Summary
Quantifying differential genome occupancy by chromatin immunoprecipitation (ChIP) remains challenging due to variation in chromatin fragmentation, immunoprecipitation efficiencies, and intertube variability. In this protocol, we add heterologous spike-ins from Drosophila chromatin as an internal control to the mice chromatin before immunoprecipitation to normalize for technical variation in ChIP-qPCR or ChIP-seq. The choice of spike-in depends on the evolutionary conservation of the protein of interest and the antibody used.
For complete details on the use and execution of this protocol, please refer to Greulich et al. (2021).
Subject areas: Sequence analysis, Cell Biology, ChIPseq, Molecular Biology, Chromatin immunoprecipitation (ChIP)
Graphical abstract

Highlights
-
•
Chromatin immunoprecipitation (ChIP) maps protein-binding sites in the genome
-
•
Heterologous spike-in ChIP reduces technical variation in ChIP experiments
-
•
Applicable to proteins with high interspecies conservation
Quantifying differential genome occupancy by chromatin immunoprecipitation (ChIP) remains challenging due to variation in chromatin fragmentation, immunoprecipitation efficiencies, and intertube variability. In this protocol, we add heterologous spike-ins from Drosophila chromatin as an internal control to the mice chromatin before immunoprecipitation to normalize for technical variation in ChIP-qPCR or ChIP-seq. The choice of spike-in depends on the evolutionary conservation of the protein of interest and the antibody used.
Before you begin
Chromatin ImmunoPrecipitation (ChIP) profiles the occupancy of DNA-associated factors within cells by either quantitative PCR (ChIP-qPCR) or with next-generation sequencing (ChIP-seq). Cells or tissues are fixed with formaldehyde and target-specific antibodies are used to precipitate the protein of interest after cell, nuclear lysis and chromatin fragmentation. Due to the complex protocol design and the variability in chromatin fragmentation between samples, inter-tube comparability is difficult to achieve without internal controls. One method to address this issue is the use of loci that do not change occupancy between samples to perform inter-tube normalization (Allhoff et al., 2016). This resembles the "housekeeping" approach used to normalize mRNA expression in standard quantitative RT-PCR of cDNAs. Such approaches are the basis of software packages like Thor (Allhoff et al., 2016), DiffBind (Ross-Innes et al., 2012) or DESeq2 (Love et al., 2014) for ChIP-seq quantification. In ChIP-qPCR, the choice of a positive locus bound by the protein of interest that does not change across conditions can be very challenging. Often, the genomic targets of the protein of interest or the behavior in response to experimental perturbations are unknown. ChIP-seq approaches are slightly more robust, as long as the majority of binding events is unaffected by the experimental perturbation. In this case, several control regions can be picked for normalization, avoiding the bias of selecting one control region. Those regions might be promoter regions of well characterized housekeeping genes, as suggested by Allhoff et al. (Allhoff et al., 2016). However, those normalization approaches are limited by the assumption that the occupancy of the protein of interest remains unaltered at the majority of sites (ChIP-seq), or at a particular locus (ChIP-qPCR), under the various conditions studied.
One example, where this assumption is violated, was reported when profiling H3 lysine 27 trimethylation (H3K27me3) after inhibition of EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit). The inhibition of EZH2, the major histone methyltransferase for H3K27, resulted in a global loss of H3K27me3. In order to quantify the observed genomic changes in H3K27me3, the authors developed “parallel ChIP”. By spiking Drosophila melanogaster chromatin into the target samples (Mus musculus), they internally controlled for inter-tube variation by performing ChIP against the Drosophila-specific histone variant H2Av (Egan et al., 2016).
Here, we describe a very similar approach by using an antibody that specifically detects the protein of interest in the samples of target as well as the spike-in species. Our method is limited to the conservation of the protein of interest, but it is independent of the variability in IP efficiencies of the antibody. We successfully used this approach for profiling several histone modifications (H3K27ac, H3K4me1, H3K4me2 and H3K4me3) by ChIP-seq and ChIP-qPCR in murine macrophages with spike-ins from Drosophila S2 cells (Greulich et al., 2021). We also performed spike-in normalization for transcription factors with limited conservation in murine cells using human spike-in chromatin (HEK293 cells) by ChIP-qPCR.
Before you begin with the actual experiment, crosslink target and spike-in cells/tissues, establish the sonication conditions for each cell type, design species-specific negative and positive control primers and confirm the specificity of the antibody in both species (see the following sections for more details). We emphasize here that optimal sonication conditions and a thoroughly tested antibody are crucial for a successful ChIP experiment.
Crosslinking of target or spike-in cells
Timing: 30 min
-
1.Before starting:
-
a.Plate 20 M cells in a 15 cm plate one day before fixation. Perform treatment of interest.
-
b.Prepare 1% formaldehyde∗ (FA) solution and 1 M glycine solutions in DPBS. (see materials and equipment).
 CRITICAL: Formaldehyde is toxic. Always use protective clothing and follow safety instructions when working with formaldehyde. Work in a fume hood and dispose of residuals according to local regulations for hazardous waste.
CRITICAL: Formaldehyde is toxic. Always use protective clothing and follow safety instructions when working with formaldehyde. Work in a fume hood and dispose of residuals according to local regulations for hazardous waste. -
c.Pre-cool DPBS.
-
a.
-
2.
Aspirate the medium, wash cells once with 10 mL DPBS. Aspirate again.
-
3.
Add 1% FA (10 mL/plate) and incubate for 15 min at 18°C–24°C.
Note: For proteins that are not contacting the DNA directly, a dual crosslinking with an additional protein-protein crosslinker like disuccinimidyl glutarate (DSG) might be required. In that case, start with a 30 min fixation with 2 mM DSG (in DPBS) at 4°C and proceed after aspiration of the DSG to the FA fixation without any wash step.
-
4.
Add 1.5 mL 1 M glycine and incubate for 5 min at 18°C–24°C. Rock back and forth gently to mix.
-
5.
Aspirate. Wash 2× with cold DPBS and harvest the cells by scraping off the dish.
-
6.
Transfer the cell suspension to a tube and keep on ice.
CRITICAL: For the spike-in cells, split the cells into two tubes with 10 M cells each to avoid freeze and thaw cycles.
-
7.
Spin the tubes for 5 min at 400 × g at 4°C. Aspirate Supernatant.
Pause point: Pellets might be stored at −80°C for up to one year.
Establish sonication conditions
Timing: 2 days
The number of sonication cycles and the amplitude of sonication needs to be optimized beforehand, for both the spike-in cells as well as the cells of interest (target). Ideally, chromatin is fragmented to 150 bp to 1.5 kb with as little energy added by sonication as possible. This step is very important, since “over-shearing” of the chromatin (bulk fragments below 200 bp) will reduce the IP efficiency by damaging the protein epitopes of interest. On the other hand, “under-shearing” (bulk fragments above 1 kb) will reduce the amount of purified DNA, due to loss of DNA during purification or size selection.
Test the sonication conditions by serial sonication of the same chromatin sample. For example, take a chromatin aliquot every two sonication cycles and perform reverse crosslinking overnight (see steps 42–47). The purified DNA is run on a 0.7% agarose gel. An example picture for a murine macrophage pellet sheared at 20 M cells/mL of shearing buffer, with the Bioruptor 300, is shown in Figure 1A and an example for Drosophila S2 cells in Figure 1B. In this case, the optimal number of cycles would be 12 for a mix of both cell types.
Figure 1.
Optimization of sonication conditions
(A) Chromatin from murine bone marrow derived macrophages (male mice aged 6–12 weeks) was sheared for 8–22 cycles at high settings (30s on/off) using the Bioruptor 300 (Diagenode). Here, 12 cycles appear optimal.
(B) Chromatin from Drosophila melanogaster S2 cells was sonicated for 6–16 cycles at high settings (30s on/off) using the Bioruptor 300. The optimal shearing conditions appear to be 10 cycles. (A+B) 20 μl of chromatin aliquots were taken during sonication after the indicated number of cycles, reverse-crosslinked, purified (see steps 42–47), and loaded onto a 0.7% agarose gel stained with peqGreen DNA dye.
Design and order of qPCR primers
Timing: 1–2 h
In order to test the specificity of the ChIP, design at least one primer pair for a region bound by the protein of interest in each the target and spike-in species (positive locus). Additionally, design one primer pair for a region not bound by the protein of interest (negative locus). For histone marks, use publicly available data from ENCODE (https://www.encodeproject.org/ (Davis et al., 2018)), MODENCODE data available for Drosophila melanogaster (http://gbrowse.modencode.org/fgb2/gbrowse/fly/), public track hubs on UCSC (https://www.genome.ucsc.edu/ (Kent et al., 2002)) or available data on the Gene Expression Omnibus (GEO) (Edgar et al., 2002). An example can be found in Figure 2A and 2B.
Note: If ChIP-Seq data for the mark or protein of interest is unavailable for the target or for the spike-in tissue, we recommend the generation of a ChIP-Seq data set before performing ChIP quantifications by qPCR.
Figure 2.
Selection and testing of species-specific PCR primers
(A) Drosophila melanogaster genome browser screen shot (http://gbrowse.modencode.org/fgb2/gbrowse/fly/) showing publicly available data for H3K4me2 ChIP-Seq at the eRF3 (also known as Elf) locus.
(B) UCSC genome browser track for H3K4me2 ChIP-Seq in murine bone marrow derived macrophages after 3 h 100 ng/mL LPS (purple, lower track) or 16 h 1 μM dexamethasone and 3 h 100 ng/mL LPS treatment (L+D, blue, upper track) (Greulich et al., 2021).
(C) ChIP-qPCR against H3K4me2 in either pure S2 cells (indicated by the fly), 25% S2 cells mixed with 75% murine macrophages treated with 100 ng/mL LPS for 3 h (marked by the fly + mouse symbol) or pure murine macrophages treated with LPS (marked by the mouse symbol). The mean of two biological replicates is plotted. Dots represent single data points, and error bars reflect the standard deviation. The color indicates the locus. (A+B) The red lines indicate the fragments amplified by PCR in C. The DNA sequence of the regions covered by the H3K4me2 signal in both species was used as input for Primer-BLAST, in order to design the primers for C (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, (Ye et al., 2012)).
Validate antibodies for specificity in target and spike-in species
Timing: 3 days
In order to confirm that the antibody indeed recognizes the protein of interest in both species and to validate the specificity of the PCR primer, perform ChIP-qPCR against the protein of interest (H3K4me2 in our case). Use the target species, the spike-in species and a mixture of target and spike-in species (10%–25% spike-in) as samples and follow the protocol below (steps 1–55). An example of the expected results are shown in Figure 2C. Here, we performed ChIP against H3K4me2 in Drosophila S2 cells, in murine bone marrow-derived macrophages treated with 100 ng/mL LPS for 3 h, and in a 1:4 mixture of Drosophila S2 cells with murine macrophages. The Drosophila-specific primers against H3K4me2 (eRF3 locus) (Figure 2A) are only enriched in the samples containing chromatin from Drosophila melanogaster. On the other hand, the mouse-specific primers against a H3K4me2-positive (Cxcl10/11, Figure 2B) and H3K4me2-negative (NegPol2) locus give a specific signal in samples containing murine chromatin. In addition, we observe a higher enrichment at the positive (Cxcl10/11) over the negative (NegPol2) locus in murine macrophages, indicating specificity of the antibody (Figure 2C).
CRITICAL: The protein of interest needs to be conserved between target and spike-in species in order to be recognized by the antibody in both species (see limitations).
Material preparations
Timing: 30 min
-
8.
Pre-cool centrifuges suitable for Eppendorf tubes to 4°C.
-
9.
Turn on Bioruptor and pre-cool the water bath.
-
10.
Prepare buffers (see materials and equipment) and aliquots of Fast IP, Shearing and Dilution, add EDTA-free proteinase inhibitors and store on ice.
-
11.
Pre-heat thermomixers to 99°C, 37°C or 56°C.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H3K4me2 | Abcam | Cat.#ab7766; RRID:AB_2560996 |
| Rabbit normal IgG control | Cell Signaling | Cat.#2729 RRID:AB_1031062 |
| Chemicals, peptides, and recombinant proteins | ||
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat.#11836170001 |
| cOmplete™ Ultra, EDTA-free Protease Inhibitor Cocktail | Roche | Cat.#5892953001 |
| DPBS | Gibco | Cat.#14190144 |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat.#28906 |
| Glycine, ReagentPlusTM, >= 99% | Sigma | Cat.#G7126 |
| Dexamethasone | Sigma | Cat.#D4902 |
| LPS E.COLI O111:B4 | Sigma | Cat.#LPS25 |
| 5 M NaCl | Sigma | Cat.#71386 |
| 1 M Tris-HCl, pH 7.5 | Invitrogen | Cat.#15567-027 |
| 1 M Tris-HCl, pH 8.0 | Gibco | Cat.#15568-025 |
| EDTA, 0.5 M sterile solution | VWR | Cat.#E177 |
| NaOAc trihydrate, pure Ph. Eur. | AppliChem | Cat.#A1370 |
| 100× Tris-EDTA buffer solution | Sigma | Cat.#T9285 |
| IGEPAL CA-630/NP40 | Sigma | Cat.#I3021 |
| Triton X-100 | AppliChem | Cat.#A1388 |
| SDS solution 20% (BioUltra for molecular biology) | Sigma | Cat.#05030 |
| Tween 20 | AppliChem | Cat.#A1389 |
| NaHCO3 Ph. Eur. | AppliChem | Cat.#A1353 |
| BSA (molecular biology grade) | Sigma | Cat.#A3294 |
| Nuclease-free water | Sigma | Cat.#W3513 |
| RNase A (DNase free) | AppliChem | Cat.#A38320050 |
| Proteinase K from Tritrachium album | Sigma | Cat.#P6556 |
| Glycerol 86% | Roth | Cat.#4043.3 |
| Dynabeads M-280 Sheep Anti-Rabbit IgG | Life Technologies | Cat.#11204D |
| peqGreen | PeqLab | Cat.#37-5010 |
| peqGOLD Universal-Agarose | VWR | Cat.#732-2789 |
| GeneRuler 100 bp DNA Ladder | Thermo Fisher Scientific | Cat.#SM0241 |
| Acetic acid 100% | Merck Millipore | Cat.#818755 |
| Sepharose Protein A/G beads | Rockland | Cat.#PAG50-00-0002 |
| Power SYBR Green Master Mix | Thermo Fisher Scientific | Cat.#4367659 |
| Agencourt AMPure XP beads | Beckman Coulter GmbH | Cat.#A63881 |
| Ethanol absolute for molecular biology | AppliChem | Cat.# A3678 |
| Critical commercial assays | ||
| MinElute PCR Purification Kit | QIAGEN | Cat.#28006 |
| Kapa HyperPrep Kit | Roche | Cat.#7962363001 |
| KAPA Library Quantification Kit Illumina-Rox Low | Roche | Cat.#7960336001 |
| High Sensitivity DNA Kit | Agilent | Cat.# 5067-4626 |
| Qubit dsDNA HS Assay Kit | Life Technologies | Cat.#Q32854 |
| Gel Cassettes, Pippin Prep, dye-free | Sage Science | Cat.#CDF2010 |
| Deposited data | ||
| ChIP-Seq in RAW264.7 | Greulich et al 2021 | GEO:GSE138017 |
| Experimental models: Cell lines | ||
| S2 cells (Drosophila) | Provided by Prof. P. Becker (LMU Munich, Germany) | RRID:CVCL_Z232 |
| RAW264.7 | ATCC | Cat.#TIB-71™ ; RRID:CVCL_0493 |
| RAW264.7 Setd1aDel/+ | Greulich et al.2021 | |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
|
Fkbp5_forward AGCGTAAGATCGCGAGAGTG |
Eurofins | N/A |
|
Fkbp5_reverse AACGTCGAGGGTGGAGAGTA |
Eurofins | N/A |
| NegPol2_forward TAGCTTTCGACAGAGGTCCTAAG |
Eurofins | N/A |
| NegPol2_reverse CCGAAGGTGGCCGGTTGT |
Eurofins | N/A |
|
eRF3_forward TGTTAACAATCACGGCGCAT |
Eurofins | N/A |
|
eRF3_reverse AAACGACACCACAAAGCGAA |
Eurofins | N/A |
|
Cxcl10/11_forward CCAGGCTATGCGATGGTTCA |
Eurofins | N/A |
|
Cxcl10/11_reverse GATAAGAGCTGACCCGGCAA |
Eurofins | N/A |
| TruSeq Illumina universal adapter AATGATACGGCGACCACCGAGATCTACACT CTTTCCCTACACGACGCTCTTCCGATC∗T |
IDT | N/A |
| TruSeq Illumina index adapter Phos/GATCGGAAGAGCACACGTCTG AACTCCAGTCACNNNNNNATCTCGT ATGCCGTCTTCTGCTTG |
IDT | N/A |
| Software and algorithms | ||
| FastQC | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 |
| Trimmomatic | Bolger et al. 2014 | RRID:SCR_011848; http://www.usadellab.org/cms/index.php?page=trimmomatic |
| BWA-MEM v0.7.13 | Li et al. 2009 | RRID:SCR_010910; https://sourceforge.net/projects/bio-bwa/files/ |
| Picard Tools v2.0.1 | http://picard.sourceforge.net/). | RRID:SCR_006525 |
| Samtools v1.8 | Li et al. 2009 | RRID:SCR_002105; http://www.htslib.org/ |
| Deeptools v3.0.2-1 | Ramirez et al. 2014 | RRID:SCR_016366; https://deeptools.readthedocs.io/en/develop/ |
| Integrated genome browser v9.0.2 | Freese et al. 2016 | RRID:SCR_011792; https://www.bioviz.org/ |
| MACS2 v2.1.1.20160309 | Zhang et al. 2008 | RRID:SCR_013291; https://github.com/macs3-project/MACS |
| BEDtools v2.25.0 | Quinlan and Hall 2010 | RRID:SCR_006646; https://bedtools.readthedocs.io/en/latest/# |
| DESeq2 v1.30.1 | Love et al. 2014 | RRID:SCR_015687; https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GenomicRanges v1.42.0 | Lawrence et al. 2013 | RRID:SCR_000025; https://www.bioconductor.org/packages/2.13/bioc/html/GenomicRanges.html |
| R v3.6.1 | Team 2017 | RRID:SCR_001905; https://cran.r-project.org/ |
| Other | ||
| Bioruptor 300 with water cooler | Diagenode | Cat. # B01060001, B02010002, B02020004 |
| QuantStudio 6 and 7 | Applied Biosystems | N/A |
| DynaMag-2 | Thermo Fisher Scientific | Cat. #12321D |
| DynaMag-PCR | Thermo Fisher Scientific | Cat. #492025 |
| Qubit 2.0 | Thermo Fisher Scientific | Cat. #Q32871 |
| Pippin Prep | Sage Science | N/A |
| NovaSeq 6000 | Illumina | N/A |
| Bioanalyzer 2010 | Agilent | N/A |
Alternatives: Any supplier may provide chemicals. Chemicals should be molecular-biology grade.
Alternatives: ChIP fragmentation with the Bioruptor might be other sonication devices like the Covaris systems (https://www.covaris.com/products-services/instruments) or probe sonicators. Alternatively, chromatin can be fragmented enzymatically using micrococcal nuclease (MNase). Either way, the optimal conditions have to be established before performing the actual ChIP experiment.
Alternatives: Any other equipment supplier may provide equivalent equipment.
Materials and equipment
Buffers
| 1% formaldehyde (FA) | Final concentration | Amount |
|---|---|---|
| Formaldehyde∗ (16%, MeOH-free) | 1% vol/vol | 1 mL |
| DPBS (1×) | 1× | 15 mL |
| Total | 16 mL |
1% formaldehyde can be stored at 4°C–8°C for up to 1 day.
| 1 M glycine | Final concentration | Amount |
|---|---|---|
| Glycine | 1 M | 75.07 g |
| DPBS (1×) | 1× | 1 l |
| Total | 1 l |
Sterilize by filtering using a 0.22-μm filter. Store 1 M glycine at 18°C–24°C for up to 1 year. Always prepare aliquots.
| Fast IP buffer | Final concentration | Amount |
|---|---|---|
| NaCl (5 M) | 150 mM | 33.3 mL |
| Tris-HCl pH=7.5 (1 M)∗ | 50 mM | 50 mL |
| EDTA pH=7.5 (0.5 M) | 5 mM | 10 mL |
| NP-40/IGEPAL CA-630 (100%) | 0.5% vol/vol | 5 mL |
| Triton X-100 (100%)∗ | 1% vol/vol | 10 mL |
| ddH2O | N/A | 891.7 mL |
| Total | 1 L |
Sterilize by filtering using a 0.22 μm filter. Store Fast IP Buffer at 4°C for up to 6 months. Always prepare aliquots.
| Shearing buffer | Final concentration | Amount |
|---|---|---|
| SDS (20%)∗ | 1% vol/vol | 50 mL |
| EDTA pH=8.0 (0.5 M) | 10 mM | 20 mL |
| Tris-HCl pH=8.0 (1 M)∗ | 50 mM | 50 mL |
| ddH2O | N/A | 880 mL |
| Total | 1 l |
Sterilize by filtering using a 0.22 μm filter. Store Shearing Buffer at 18°C–24°C for up to 6 months. Always prepare aliquots.
Alternatives: If the SDS concentration is too high for a particular antibody (Troubleshooting 7), low-SDS shearing buffers with SDS contents as low as 0.1% can be tried. Note that sonication conditions need to be re-established when changing the shearing buffer. We have observed prolonged sonication times (Bioruptor) to be required when reducing the SDS content.
Alternatives: Add 0.1% Triton X-100 to avoid SDS precipitation while shearing especially if longer sonication times are required.
| Dilution buffer | Final concentration | Amount |
|---|---|---|
| SDS (20%)∗ | 0.01% vol/vol | 0.5 mL |
| Triton X-100 (100%)∗ | 1.1% vol/vol | 11 mL |
| EDTA pH=8.0 (0.5 M) | 1.2 mM | 2.4 mL |
| Tris-HCl pH=8.0 (1 M)∗ | 16.7 mM | 16.7 mL |
| NaCl (5 M) | 167 mM | 33.4 mL |
| ddH2O | N/A | 936 mL |
| Total | 1 l |
Sterilize using a 0.22 μm filter. Store Dilution Buffer at 4°C for up to 6 months. Always prepare aliquots.
| Library elution buffer (LEB) | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH=8.0 (1 M)∗ | 10 mM | 100 μL |
| Tween-20 (100%) | 0.05% vol/vol | 5 μL |
| Nuclease-free water | N/A | 9.9 mL |
| Total | 10 mL |
Store LEB buffer at 18°C–24°C for up to 2 days.
| 1× TE buffer | Final concentration | Amount |
|---|---|---|
| 100× TE (0.2 μm-filtered, 1 M Tris and 100 mM EDTA) | 1× (10 mM Tris and 1 mM EDTA) | 100 μl |
| Nuclease-free water | N/A | 9.9 mL |
| Total | 10 mL |
Store TE buffer at 18°C–24°C for up to 1 month.
| 3 M NaOAc pH=5.2 | Final concentration | Amount |
|---|---|---|
| NaOAc (molecular biology-grade, anhydrous) | 3 M | 82.03 g |
| Nuclease-free water | N/A | 1 l |
| Total | 1 l |
Adjust pH to 5.2 with acetic acid∗ and filter with a 0.22 μm filter. Store NaOAc at 18°C–24°C for up to 1 year.
| 10 mg/mL RNase A stock | Final concentration | Amount |
|---|---|---|
| RNase (DNase-free) | 10 mg/mL | 50 mg |
| 1× TE buffer | 1× | 5 mL |
| Total | 5 mL |
Aliquot and store RNase A at −20°C for up to one year. Aliquots might be thawed for up to 5 times.
| 10 mg/mL Proteinase K stock | Final concentration | Amount |
|---|---|---|
| Proteinase K | 10 mg/mL | 100 mg |
| Nuclease-free water | N/A | 10 mL |
| Total | 10 mL |
Aliquot and store proteinase K at −20°C for up to one year. Aliquots might be thawed for up to 5 times.
| 5% BSA | Final concentration | Amount |
|---|---|---|
| BSA | 5% w/v | 2.5 g |
| Nuclease-free water | N/A | 50 mL |
| Total | 50 mL |
Filter the buffer with a 0.45 μm syringe filter. Aliquot and store 5% BSA at −20°C for up to one year. Aliquots might be thawed for up to 5 times.
| 6× color-less loading dye | Final concentration | Amount |
|---|---|---|
| Glycerol (89%) | 30% vol/vol | 340 μl |
| Nuclease-free water | N/A | 660 μl |
| Total | 1 mL |
The loading dye is stable at 8°C for 6 months.
| 1 M NaHCO3 | Final concentration | Amount |
|---|---|---|
| NaHCO3 (molecular biology-grade) | 1 M | 84 mg |
| Nuclease-free water | N/A | 1 mL |
| Total | 1 mL |
1 M NaHCO3 needs to be prepared freshly and only lasts for 2–3 h at 18°C–24°C.
| Bead elution buffer | Final concentration | Amount |
|---|---|---|
| fresh NaHCO3 (1M) | 100 mM | 100 μl |
| SDS∗ (20%) | 1% vol/vol | 50 μl |
| Nuclease-free water | 850 μl | |
| Total | 1 mL |
Bead Elution Buffer needs to be prepared freshly and is stable at 18°C–24°C for 2–3 h.
| 50× TAE buffer | Final concentration | Amount |
|---|---|---|
| Tris base | 2 M | 484.0 g |
| Acetic Acid∗ (100%) | 1 M | 114.2 mL |
| EDTA (di-sodium salt) | 0.05 M | 37.2 g |
| Deionized H20 | N/A | Fill to 2 l |
| Total | 2 l |
The pH of the buffer should be 8.3 and it can be stored at 18°C–24°C for 6 month.
| 1× TAE buffer | Final concentration | Amount |
|---|---|---|
| 50× TAE buffer | 1× | 100 mL |
| Deionized H20 | N/A | 4.99 l |
| Total | 5 l |
The buffer can be stored at 18°C–24°C for 6 month.
Note: For the Fast IP, Shearing and Dilution Buffers, prepare aliquots before starting the experiment and add EDTA-free proteinase inhibitors freshly. These solutions are stable for 1 day at 4°C.
CRITICAL: Harmful chemicals are indicated with an asterisk (∗) here or in the following protocol.
Formaldehyde
Flammable liquid and vapor. Harmful if swallowed. Causes severe skin burns and eye damage. May cause an allergic skin reaction. Causes serious eye damage. Toxic if inhaled. May cause cancer (inhalation). Toxic to aquatic life. Only work with formaldehyde when specifically instructed. Keep away from heat sources and open flames. Take precautionary measures against static discharge. Do not eat, drink or smoke when using formaldehyde. Avoid environmental release. Wear protective clothing, protective gloves, eye and face protection. Wash contaminated clothing before reuse. Store in well-ventilated cabinets and keep containers tightly closed. Dispose of content and containers to comply with local regulatory authorities.
Ethanol
Highly flammable liquid and vapor. Causes serious eye damage/irritation. Keep Ethanol away from heat sources, open flames. Do not smoke. Keep containers tightly closed. Large amounts should be stored in fireproof cabinets.
Acetic acid
Flammable liquid and vapor. Harmful if inhaled. Causes severe skin burns and eye damage. Causes serious eye damage. Harmful to aquatic life. Keep away from heat sources. Store in fireproof cabinets. Take precaution measures against static discharge. Wash exposed skin thoroughly after handling. Only work in well-ventilated areas or under the fume hood. Wear protective clothing, protective gloves, eye and face protection.
SDS
Flammable solid. Harmful if swallowed or inhaled, causes skin irritation or serious eye damage. May cause respiratory irritation. Harmful to aquatic life with long lasting effects. Keep away from heat sources. Avoid breathing dust by using pellets or masks. Wear protective clothing and eye protection.
Tris-HCl
May cause eye and skin irritation. Wear protective clothing.
Triton X-100
Harmful if swallowed. Causes serious eye damage. Toxic to aquatic life with long lasting effects. Do not eat, drink or smoke when using Triton X-100. Avoid environmental release. Wear protective gloves/eye protection.
RNase A
May cause allergy or asthma symptoms or breathing difficulties if inhaled. Avoid breathing dust/vapor.
Proteinase K
Causes skin irritation. May cause allergic skin reaction. Causes serious eye irritation. May cause allergy or asthma symptoms or breathing difficulties if inhaled. May cause respiratory irritation. Do not breathe dust. Wear protective gloves/eye protection.
Step-by-step method details
Cell lysis
Timing: 30 min
In this step, the frozen, formaldehyde-fixed pellets from target and spike-in cells are processed for cell lysis. All steps are performed on ice.
Note: Target and spike-in cells are processed separately to allow for alternative cell lysis protocols e.g., when working with tissues instead of cell lines.
Alternatives: If the cell lysis and sonication conditions for target and spike-in cells are identical, both might already be mixed in step 2.
-
1.
Thaw target and spike-in cell pellets on ice.
Note: For one ChIP-seq experiment targeting a transcription factor, use 40 M cells, and for histone marks, use 20 M cells. For ChIP-qPCR, 2 M cells per IP are recommended. 5% of the experimental cell number is required for spike-in per ChIP-seq experiment, and 15%–25% should be used for ChIP-qPCR. Use spike-in pellets accordingly.
CRITICAL: The amount of spike-in chromatin must be adjusted depending on the genome size differences between the target and spike-in species and according to the genomic coverage of the protein of interest. The above-mentioned amount of spike-in chromatin is optimized for murine cells combined with Drosophila spike-in, and for performing ChIP-Seq against widely occurring histone marks such as H3K4me1/me2/me3 or H3K27ac. See Troubleshooting 5.
-
2.
Resuspend 20 M cells in 1 mL ice-cold FastIP buffer. Let the chromatin sit on ice for 10 min. During this time, pull the cell suspension through an insulin syringe once for mechanical cell lysis.
-
3.
Spin at 12,000 × g for 1 min at 4°C. Aspirate the supernatant using either vacuum or a pipette. Leave the pellet untouched. Some liquid may remain.
-
4.
Repeat steps 2 and 3.
CRITICAL: Cell and tissue lysis needs to be optimized individually for each tissue or cell line. We refer to Mir at al. (Mir et al., 2019) for recommendations.
Spike-in and sonication
Timing: 30 min per 3 samples (depending on the Bioruptor tube holder)
In this step, 5% of spike-in chromatin is mixed with the target chromatin and fragmented by sonication.
-
5.
Resuspend cell pellets completely in 1 mL ice-cold Shearing Buffer per 20 M cells.
CRITICAL: The Shearing Buffer needs to be cold but not “cloudy”. In case of SDS precipitation, warm the sample briefly and vortex it to solubilize all precipitates.
-
6.
Shortly before distributing the spike-in chromatin, mix spike-in nuclei very well and add 5% of spike-in per target chromatin per 1.5 mL Bioruptor TPP tube.
Note: For sonication in 1.5 mL Bioruptor TPP tubes, do not fill the tube with more than 550 μl of chromatin, to ensure a complete immersion of the liquid-filled tube in the water bath. If 20 M target cells are resuspended in 1 mL Shearing Buffer and 10 M spike-in cells in 500 μl Shearing Buffer, aliquot 25 μl spike-in nuclei per 1.5 mL Bioruptor tube and add 500 μl of target chromatin. Two Bioruptor tubes per 20 M target cells are required during sonication.
Troubleshooting 1: Different sonication conditions for target and spike-in cells or tissues.
-
7.
Continue with sonication using a Bioruptor 300 or similar model (Diagenode).
Note: If the Bioruptor tube holder cannot fit all the tubes in one round, distribute aliquots from the same sample across different rounds to avoid sonication biases introduced by the samples being in different sonication rounds.
-
8.
Unite the sheared chromatin from each sample aliquot inside a 2 mL low binding Eppendorf tube and store on ice until proper sonication is confirmed by fast-reverse cross-linking.
Fast reverse cross-linking
Timing: 2–3 h
Here, the appropriate size of the chromatin fragments is validated before proceeding with immunoprecipitation.
-
9.
Take 20 μL of sheared chromatin from each sample (e.g., experimental condition) and adjust the volume to 100 μL with 1×TE buffer.
-
10.
Add 4 μL 5 M NaCl and incubate at 99°C for 15 min mixing at 1,000 rpm in a thermomixer.
-
11.
Remove samples from the thermomixer and wait for them to cool down to 18°C–24°C.
-
12.
Add 0.5 μL RNase A∗ (10 mg/mL stock). Incubate for 15 min at 37°C and 300 rpm.
-
13.
Add 4 μL 1 M Tris-HCl∗ pH=7.5, 2 μL 0.5 M EDTA pH=8 and 1 μL Proteinase K∗ (10 mg/mL stock). Incubate at 56°C for 1 h, while mixing at 300 rpm.
-
14.
Purify the samples with the MinElute PCR purification kit (QIAGEN) following the manufacturer's instructions. Elute the samples in 15 μL elution buffer.
Note: Adjust the pH of the PB buffer if the color is not yellow, by adding 1 μl 3 M NaOAc pH=5.2.
-
15.
Mix each sample with 4 μl of 6× colorless DNA loading dye and load onto a 0.7% agarose gel containing peqGreen or similar.
Note: Dyes in the sample loading buffer might lower the fluorescence intensity of the sample DNA at the front of the dye and thereby affect the visibility of the fragmented chromatin.
-
16.
If required, add additional sonication cycles and repeat the fast reverse crosslink.
CRITICAL: The fragment size of the chromatin should be comparable between all samples under study. See Figure 3 for an example.
Figure 3.
DNA fragment size verification
Purified DNA form macrophages treated with LPS (A) or LPS plus Dexamethasone (B) was loaded onto a 0.7% agarose gel stained with peqGreen DNA dye. Each sample contains 5% spike-in chromatin from Drosophila S2 cells
Immunoprecipitation
Timing: 2 days
In this step, the chromatin fraction bound by the factor of interest is purified using immunoprecipitation (IP).
-
17.
Leave the fragmented chromatin at 18°C–24°C until all SDS precipitates are dissolved. Occasionally vortex samples gently. (1–2 min)
-
18.
Centrifuge at 12,000 × g for 10 min at 4°C.
-
19.
Take the top 90% of each sample’s supernatant (900 μl for one 20 M cell pellet) into a 15 mL conical tube. Do not touch the pellet (cell debris).
-
20.
Transfer 9 μl of supernatant from each sample to a 1.5 mL tube as an input control (1% input) and freeze at −20°C until de-crosslinking in step 41. Discard the remaining volume and the pellet.
Note: The amount of input material might be increased to 10%, if an independent input sample is required for sequencing. Adjust the percentage input calculations accordingly.
-
21.
Add 8.1 mL Dilution Buffer to the 15 mL conical tube containing the sample (1:10 dilution of the sheared chromatin).
Note: For ChIP-qPCR, take 10% input (9 μl) and 90 μl of chromatin per IP. Dilute the chromatin with 810 μl of dilution buffer and perform IPs in 1.5 mL low binding Eppendorf tubes. Adjust the percentage input calculations accordingly.
Alternatives: If the epitope of interest is expressed at low levels or if the antibody affinity is low, reducing the dilution of the sheared chromatin might be advantageous. In this case, reduce the amount of SDS in the shearing buffer (see Troubleshooting 7).
Alternatives: An additional pre-clearing step might be included, in the event that high background signal is detected (see Troubleshooting 7).
-
22.
Add 4 μg of antibody to each 15 mL conical tube containing the diluted sample.
Note: For ChIP-qPCR, use 1 μg of antibody for each IP.
Note: Antibody concentrations need optimization and depend on the antibody affinity and the amount of epitope present within the sample.
Note: Antibodies must be tested for specificity. See Troubleshooting 2.
-
23.
Rotate tubes 12–16 h at 4°C.
-
24.Block Dynabeads.Note: The type of beads depends on the isotype and species of the ChIP antibody. For polyclonal antibodies raised in rabbit, we recommend sheep anti-rabbit IgG Dynabeads (Life Technologies).
-
a.Transfer 60 μL of well suspended Dynabeads/IP from the stock bottle into a 1.5 mL low binding tube and place the tube into a magnetic rack.
-
b.Add 1 mL Dilution Buffer. Remove the tube from the rack and resuspend the beads by inverting and flicking the tube.
-
c.Place the tube back into the magnetic rack and wait for 30 s until the liquid is cleared from the beads.
-
d.Aspirate the supernatant without disturbing the beads.
-
e.Repeat b-d twice more.
-
f.Add 1 mL Dilution Buffer supplemented with 0.5% BSA (molecular biology-grade), completely resuspend the beads by flicking the tube and incubate 12–16 h at 4°C while rotating slowly.
-
a.
Note: For ChIP-qPCR, use 20 μL of sepharose protein A/G beads per IP. Instead of the magnetic rack, spin the beads 30 s at 300 × g for washing. Take care to never vortex sepharose beads.
Alternatives: Dynabeads perform equally well in ChIP-qPCR.
-
25.
The next day, spin the 15 mL conical tubes containing the IP for 20 min at 3,600 × g at 4°C.
Note: For ChIP-qPCR, spin IPs at 12,000 × g for 10 min at 4°C (1.5 mL Eppendorf tubes).
-
26.During the centrifugation, wash the blocked beads 3 times in Dilution Buffer.
-
a.Place the Dynabeads in the magnetic rack. Wait 30 s until all beads are collected and the liquid is cleared.
-
b.Aspirate the supernatant carefully without losing beads.
-
c.Add 1 mL Dilution Buffer, remove the tube from the magnetic rack and resuspend the beads by flicking the tube.
-
d.Place the Dynabeads inside the magnetic rack. Wait 30 s until all the beads are collected and the liquid cleared, and aspirate the supernatant carefully.
-
e.Repeat c-d twice more.
-
f.Resuspend beads in 65 μl Dilution buffer/IP.
-
a.
Note: For sepharose beads, spin the beads 30 s at 300 × g and 4°C instead of using the magnetic rack.
-
27.
Aliquot 15 μL Dynabeads into fresh 2 mL low binding Eppendorf tubes (4 tubes for one 20 M cell pellet).
-
28.
After centrifugation, take the top 90% (approx. 8 mL) of chromatin. On ice, aliquot 2 mL per 2 mL low binding Eppendorf tube containing Dynabeads beads (from step 27).
Note: For ChIP-qPCR, transfer the top 90% of the chromatin (800 μl) into a new 1.5 mL low binding tube that contains 20 μL of sepharose beads in Dilution Buffer.
-
29.
Incubate for 6 h at 4°C while slowly rotating the tubes.
-
30.Note: For ChIP-qPCR, incubate for 3 h at 4°C while rotating. On ice, wash the beads with the immobilized chromatin of interest with FAST IP Buffer. Place magnetic racks on ice.
-
a.Place the 2 mL Eppendorf tubes inside the magnetic rack. Wait for 30 s until the liquid is cleared.
-
b.Aspirate the supernatant and add 1 mL ice-cold Fast IP buffer.
-
c.Take the tubes from the magnetic rack and mix by inverting and flicking the tubes. Let the samples sit on ice for 2 min.
-
d.Repeat a-c four more times.
-
a.
Note: For sepharose beads, spin beads 30 s at 300 × g and 4°C instead of using the magnetic rack.
-
31.
During the washes, prepare the Bead Elution Buffer.
Note: Steps 32 and 33 are only required for ChIP-seq samples
-
32.
After the fifth wash, add 100 μL ice-cold 1× TE buffer to each tube. Unite the beads from the four 2 mL tubes (for one 20 M cell pellet) by removing the tubes from the magnetic rack and resuspending the beads with the P200. Collect all beads from one sample in one tube. Afterward, one tube with 400 μl bead suspension per sample remains.
-
33.
Add another 50 μL ice-cold 1× TE buffer to each one of the empty tubes, resuspend any remaining beads and unite with the beads from step 32 (650 μl beads in 1×TE).
-
34.
Place the tubes into the magnetic rack, wait 1 min until the solution is cleared, and carefully remove the supernatant with a pipette. Be careful, the beads will only be loosely attached in TE buffer.
Note: For ChIP-qPCR, the steps 32 and 33 are not required, since every tube is a separate IP.
-
35.
Add 1 mL 1× ice-cold TE buffer, resuspend the beads by inversion and by flicking the tubes, and place them back into the magnetic rack. Wait 1 min until the solution is cleared.
-
36.
Carefully pipet the TE buffer off using a pipette. Shortly spin the beads and transfer the tube back to the magnetic rack. Remove any remaining supernatant.
Note: For sepharose beads, spin beads 30 s at 300 × g and 18°C–24°C instead of using the magnetic rack.
-
37.
Add 100 μL Bead Elution Buffer per tube, vortex and incubate 15 min at 18°C–24°C and 1,000 rpm in a thermomixer.
Note: For ChIP-qPCR, add 50 μL Bead Elution Buffer to each IP.
-
38.
Place the tubes into the magnetic rack and collect the supernatant into fresh 1.5 mL low binding Eppendorf tubes.
-
39.
Add another 100 μL Bead Elution Buffer to the beads, vortex and incubate again inside the thermomixer for 15 min at 18°C–24°C and 1,000 rpm.
Note: For ChIP-qPCR, add 50 μL bead elution buffer and spin beads 30 s at 300 × g and 18°C–24°C instead of using the magnetic rack.
-
40.
Shortly spin the tubes in a bench top centrifuge. Place the tubes into the magnetic rack and collect the supernatant into the 1.5 mL tubes from step 38. The final volume of the eluted chromatin is 200 μL.
Note: For ChIP-qPCR, the final volume should be 100 μL.
-
41.
Thaw the input sample and adjust the volume of the input sample to 200 μL using 1× TE buffer.
-
42.
Add 8 μL of 5 M NaCl to the 200 μL eluted chromatin and input, vortex and heat at 65°C for 12–16 h. Do not shake.
Note: For ChIP-qPCR, adjust the volume of the input sample to 100 μL and add 4 μL of 5 M NaCl.
DNA purification
Timing: 3 h
In this step, the chromatin is de-crosslinked and the DNA is purified.
-
43.
Add 1 μL of RNase A∗ (10 mg/mL stock) to the eluted DNA and input samples.
-
44.
Incubate for 30 min at 37°C and 300 rpm in a thermomixer.
-
45.
Add 4 μL 0.5 M EDTA pH=8, 8 μL 1 M Tris-HCl∗ pH=7.5 and 1 μL Proteinase K∗ (10 mg/mL stock).
-
46.
Incubate in a thermomixer for 2 h at 56°C and 300 rpm.
-
47.Purify the de-crosslinked chromatin using the MinElute PCR purification kit from QIAGEN.
-
a.Add 1,110 μL PB buffer and 50 μL 3M NaOAc pH=5.2. The color of the pH indicator should be yellow.Note: For ChIP-qPCR, add 500 μL PB buffer and 20 μL 3M NaOAc pH 5.2.
-
b.Mix with a pipette and load 700 μl de-crosslinked chromatin in PB buffer onto the spin column and spin at 20,000 × g for 30 s at 18°C–24°C.
-
c.Discard the flow-through.
-
d.Repeat step b with the remaining chromatin.
-
e.Wash with 750 μL PE buffer (containing EtOH∗) to the spin column.
-
f.Spin at 20,000 × g for 30 s and discard the flow- through.
-
g.Spin the empty column at 20,000 × g for 1 min to remove residual PE buffer.
-
h.Add 16 μL EB buffer and spin at 20,000 × g for 1 min.
-
a.
Pause point: For ChIP-seq, the purified ChIP DNA might be stored at −20°C until libraries can be prepared. Before freezing the samples, take a 2 μl aliquot for qPCR (see steps 48–55)) and another 1–2 μl aliquot to measure the DNA concentration with Qubit. Avoid freeze and thaw cycles.
CRITICAL: Purified DNA for ChIP-qPCR is very unstable due to its low concentration. ChIP samples with lowly concentrated DNA tend to lose a higher fraction of DNA to adsorption by the tube walls and subsequent denaturation of smaller fragments (Gaillard, 1998; Zhong et al., 2017). Low-concentrated ChIP DNA (as from ChIP-qPCR) should be processed immediately. The maximal storage time is 12–16 h at −20°C.
Quantitative PCR (qPCR)
Timing: 3 h
In this step, the enrichment is quantified. It serves as a quality control of ChIP samples designated for sequencing.
-
48.
Thaw SYBR Green PCR master mix and prepare 10-μM dilutions of the primers for at least one negative and positive locus.
-
49.
Take 2 μl from the eluted ChIP or input DNA, and add 118 μl of nuclease-free water (1:60 dilution). This is sufficient for 8 qPCRs in a 384-well format when performing triplicates.
Note: For ChIP-qPCR experiments, only dilute the eluted DNA (ChIP or input) 1:15 at the highest, depending on how many qPCRs will be performed. Further dilution will make the spike-ins undetectable.
-
50.
Create five standards by making serial dilutions of the input samples, as indicated in the following table:
| Standard | Dilution |
|---|---|
| Std1 | 1:2 from all diluted inputs (step 49) |
| Std2 | 1:10 from Std1 |
| Std3 | 1:10 from Std2 |
| Std4 | 1:10 from Std3 |
| Std5 | 1:10 from Sdt4 |
-
51.
Further dilute the input 1:5.
-
52.
Add 4.5 μl of each standard, water (as non-template control), input and ChIP samples to 3 of the 384 wells each (triplicates).
-
53.
Prepare the qPCR master mix based on the following table. Replace n by the number of ChIP samples (including all input samples).
| qPCR master mix | |||
|---|---|---|---|
| Reagent | Final concentration | Amount per reaction | Amount |
| SYBR Green PCR master mix (2×) | 1× | 5 μL | |
| Forward primer (10 μM) | 0.25 μM | 0.25 μL | |
| Reverse primer (10 μM) | 0.25 μM | 0.25 μL | |
| Total | 5.5 μL | ||
-
54.
Run the following protocol on the qPCR machine:
| qPCR cycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial activation/denaturation | 95°C | 10 min | 1 |
| Denaturation | 95°C | 15 sec | 45× |
| Annealing/extension/data acquisition | 60°C | 1 min | |
Optional: Run a melt curve analysis.
-
55.
Confirm that the reaction efficiency is between 90%–110% by analyzing the standard curve.
Quality control
Before proceeding to library preparation, the following two quality control standards must be met.
-
56.
Specific enrichment for the factor of interest, as determined by ChIP-qPCR.
Test the enriched chromatin for a positive locus occupied by the factor of interest and for a negative locus that should not be bound by the factor of interest (see Figure 6).
Note: The negative control is of utmost importance, as ChIP-seq experiments are not usually compared to IgG controls to test whether an antibody is specific.
-
57.
The majority of the input chromatin should be fragmented between 200 and 1500 bp as determined by agarose gel electrophoresis (see Figures 1 and 3).
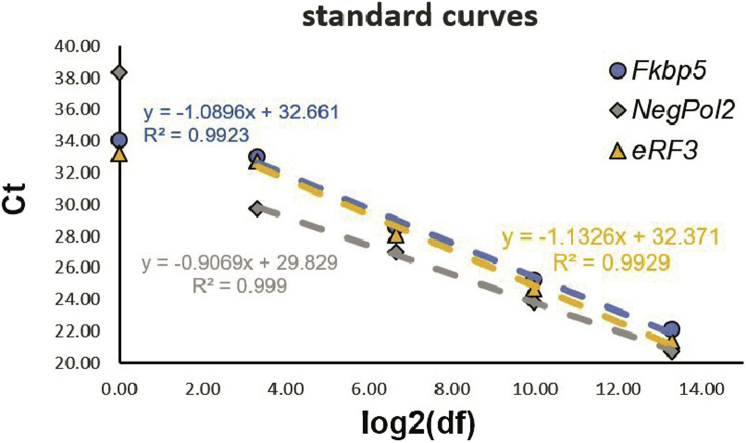
Figure 6.
Standard curve for Fkbp5 (circle, blue), NegPol2 (rectangle, gray) and eRF3 (triangle, orange) for ChIP-qPCR in murine macrophages with Drosophila S2 cell spike-in
The mean Ct values of three technical replicates are plotted as data points, and the linear regression for S1-S4 is presented as dashed line. The linear regression formulas and regression coefficients R2 are given.
Library preparation
Timing: 2 days for 1 to 20 libraries
Timing: 3 h until next pause point per five libraries for steps 66and 67
Timing: approx. 3–16 h for step 68
Timing: 2 h depending on the cycle number for steps69and 70
Timing: 3 h for 1–10 libraries for step 71
In this part, the ChIP DNA is prepared for sequencing on an Illumina NGS machine using a ligation-based approach (see Figure 4).
-
58.
Bring AMPure XP beads to 18°C–24°C.
-
59.
Prepare fresh 80% EtOH∗ (molecular biology-grade) with nuclease-free water.
-
60.
Prepare fresh Library Elution Buffer (LEB).
-
61.
Quantify the ChIP and input DNA using Qubit.
Note: Input DNA has to be diluted at least 1:100 for Qubit quantification.
-
62.
Dilute up to 5 ng of ChIP DNA in 50 μl nuclease-free water.
Note: If the ChIP DNA is undetectable, it might still be processed for library preparation (see Troubleshooting 3).
-
63.Perform end repair and A-tailing
-
a.Prepare one end repair reaction per sample in a PCR tube as follows:
End repair reaction
Reagent Amount Diluted ChIP DNA 50 μL End Repair & A-Tailing Buffer 7 μL End Repair & A-Tailing Enzyme Mix 3 μL Total 60 μL Buffers and enzymes are from the KAPA HyperPrep Kit. -
b.Mix thoroughly and spin samples using a benchtop centrifuge.
-
c.Incubate the samples in a thermocycler with the following program, and proceed to the next step immediately:
PCR cycling conditions
Steps Temperature Time Cycles End repair, A-tailing, 5′ phosphorylation 20°C 30 min 1 Heat inactivation 65°C 30 min 1 Hold 4°C max. 2 h
-
a.
-
64.Adapter ligation (single index)
-
a.Add the following reagents to the end-repaired and A-tailed ChIP DNA:
Ligation reaction
Reagent Amount Note End repair & A-tailing reaction product 60 μL Nuclease-free water 5 μL Prepare Master mix Ligation buffer 30 μL DNA ligase 10 μL 3 μM Adapters 5 μL Total 110 μl Buffers and enzymes are from the KAPA HyperPrep Kit. -
b.Mix thoroughly and spin samples shortly in a benchtop centrifuge.
-
c.Incubate at 20°C for 15 min in a thermocycler and proceed to the next step immediately.
-
a.
Note: The adapter identifies each sample. Accordingly, use different adapters for each sample and record the adapters used, to enable computational demultiplexing after sequencing. An adapter contains the primer binding sequence for the universal amplification primers in step 69, and a unique barcode (index) on one (single index) or both strands (dual index), which are required for the identification of each sample (see Figure 4). Adapters might be ordered from any oligonucleotide vendor.
Figure 4.
Library preparation
(A) Sequence of adapters (single index) used in our protocol. Red nucleotides mark the barcode/index sequence, purple and blue colors show the primer and the primer-binding site for each of the primers used during library amplification (step 69). Those lie within the universal part of the adapter.
(B) Schematic view of the four major steps of library preparation, including end-repair, A-tailing, adapter ligation, size selection and library amplification.
We are using single index adapters here. However, dual indices are recommended for sequencing on Illumina NovaSeq 6000 machines to avoid index hopping.
-
65.Post-ligation Clean-up (0.8× AMPure XP bead clean-up)
 CRITICAL: AMPure XP beads should be equilibrated to 18°C–24°C.Note: Timing during AMPure XP bead clean-up is important, we therefore recommend to process at most 10 libraries at the same time. Multiple rounds of 10 purifications are possible.
CRITICAL: AMPure XP beads should be equilibrated to 18°C–24°C.Note: Timing during AMPure XP bead clean-up is important, we therefore recommend to process at most 10 libraries at the same time. Multiple rounds of 10 purifications are possible.-
a.Mix AMPure XP beads well and add 88 μl per 110 μl ligation reaction using a pipette. Mix reaction by pipetting up and down for 10×.
-
b.Incubate 5 min at 18°C–24°C.
-
c.Place PCR tubes in magnetic tube holder (8-stripe) and wait until the liquid is clear (1–3 min).
-
d.Remove 175 μl of the supernatant with a pipette and discard it.
 CRITICAL: Do not disturb the beads.
CRITICAL: Do not disturb the beads. -
e.Add 200 μl of freshly prepared 80% EtOH∗ without disturbing the beads. Incubate 1 min.
-
f.Remove all EtOH∗ with a pipet without disturbing the beads.
-
g.Dry beads at 18°C–24°C (2–5 min).
 CRITICAL: Check each tube individually. Do not overdry beads. Overdried beads will show cracks.
CRITICAL: Check each tube individually. Do not overdry beads. Overdried beads will show cracks. -
h.Add 25 μl Library Elution Buffer to the beads, take the PCR tubes from the magnetic rack and pipet up and down for 10× times. Incubate 2 min at 18°C–24°C.
-
i.Place PCR tubes into the magnetic tube holder and wait until the liquid is cleared (1–5 min).
-
j.Transfer 22 μl of the supernatant to a fresh PCR tube.
-
a.
Pause point: Adapter-ligated DNA can be stored at 4°C for one week or at −20°C for one month.
-
66.Size Selection with the Pippin PrepAlternatives: Double-sided size selection with AMPure XP beads might be performed, but does not yield size ranges as accurately as the Pippin Prep. For a target range of 200–660 bp, 0.5× AMPure XP beads are added to the sample in the first step. Larger fragments are bound to the beads, while the desired fragments remain in the supernatant. New beads are added to the supernatant according to the following formula, using a left-sided ratio of 0.8×:The desired fragments are bound by the beads and small fragments are discarded together with the supernatant. The procedure is the same as described in point 65 after adjusting for the AMPure XP bead ratio.Note: With one Pippin Gel Cassette, size selection of up to five libraries can be performed. For more libraries, consecutive runs of Size Selection might be performed.
-
a.Turn the Pippin Prep on and create the following protocol:
-
i.Select the correct cassette from the drop-down menu: “2% Marker E”
-
ii.Select Range (turns orange).
-
iii.Enter 200 into the “BP Start” and 600 in the “BP End” field.
-
iv.Enter the sample ID.
-
v.Press “Use Internal Standards”, which will auto-fill the Reference Lane box.
-
vi.“End Run when Elution is Completed”
-
vii.Save the protocol.
-
i.
-
b.Calibrate the Pippin Prep
-
i.Press “Calibrate” on the control panel.
-
ii.Put the calibration fixture onto the optical nest (dark-side down).
-
iii.Close the lid and press “Calibrate”.
-
iv.After calibration, press “EXIT” to return to the main menu.
-
i.
-
c.Bring loading solution/marker mix to 18°C−24°C.
-
d.Add 8 μl of library elution buffer and 10 μl of loading solution/marker mix to each sample.
-
e.Mix thoroughly by vortexing and spin tubes in a benchtop centrifuge.
-
f.Unpack one 2% Dye-free Pippin Gel Cassette from the box and inspect it for cracks in the gel or for missing buffer (e.g., dried wells).Note: Do not use cassettes with cracks or dried wells.
-
g.Check for air bubbles in the detection regions and behind the elution wells. Dislodge them by slightly tapping against the cassette.Note: If an air bubble is visible between the plastic and the agarose, do not use this well.
-
h.Place the cassette into the optical nest of the Pippin Prep and remove the adhesive strips.
-
i.Refill buffer wells with less than 50% buffer.
-
j.Remove all buffer (approx. 50 μl) from the elution wells and replace with 40 μl fresh Electrophoresis Buffer.Note: Carefully place the pipet tip at the bottom of the elution well for refill and move upwards with the buffer level to avoid the introduction of air bubbles.
-
k.Seal elution wells with the provided adhesive tape strips.
-
l.Fill sample wells with Electrophoresis Buffer (approx. 70 μl in total).
-
m.Run the Continuity test by pressing “TEST”.Note: If a separation lane fails the continuity test, check the sample well’s buffer level. If the buffer is sufficient but the lane still fails, do not use this lane. If the elution channel failed, replace the elution buffer and rerun the continuity test. If it fails again, do not use this lane for any sample.
-
n.Fill up sample wells with Electrophoresis Buffer if required.
-
o.Remove 40 μl of Electrophoresis Buffer from each sample well that will be used.
-
p.Load 40 μl of sample into each well.Note: Place the pipet tip just below the buffer level and follow the liquid level while loading the sample to avoid air bubbles.
-
q.Select the prepared protocol and press “START”. One run takes approx. 2.5 h.
-
r.After the run, collect the samples by removing the adhesive tape from the top of the elution wells and carefully pipet 40 μl of eluted DNA into a fresh PCR tube using a pipette.
-
a.
-
67.Concentrate the size-selected library with AMPure XP beads
-
a.Combine 72 μl of well-mixed AMPure XP beads with 40 μl of size-selected ChIP DNA and mix with a pipet (10 times).
-
b.Proceed with the AMPure XP bead purification as described in points 66b-g.
-
c.Add 26 μl Library Elution Buffer to the beads, take the PCR tubes from the magnetic rack and pipet up and down 10 times. Incubate for 2 min at 18°C–24°C.
-
d.Place PCR tubes into the magnetic tube holder and wait until the liquid is cleared (1–5 min).
-
e.Transfer 24 μl of the supernatant to a fresh PCR tube.
-
a.
Pause point: ChIP DNA can be stored at 4°C for up to 24 h after size selection. Do not freeze.
-
68.
Kapa Library Quantification
Library quantification estimates the amount of adapter-ligated ChIP DNA and is required to estimate the number of amplification cycles for the library.-
a.Prepare serial dilutions of the ChIP DNA in Library Elution Buffer (LEB) as follows:
Serial dilution of ChIP DNA
Dilution Volume Volume LEB 1:500 1 μl ChIP DNA 499 μl 1:1000 100 μl 1:500 dilution 100 μl 1:2000 100 μl 1:1000 dilution 100 μl -
b.Load 4 μl of each standard (provided by the KAPA Library Quantification Kit) and sample into a 384-well plate. Perform assay in triplicates.Note: The standard concentrations are as follows: Std 1: 20 pM; Std2: 2 pM; Std 3: 0.2 pM; Std 4: 0.02 pM; Std 5: 0.002 pM; Std 6: 0.0002 pM
-
c.Add 6 μl of Kapa SYBR Green master mix per well.Note: Premix 5 mL KAPA SYBR Fast qPCR Master Mix with 1 mL of 10× Primer before the first use and prepare aliquots. All reagents are provided by the KAPA Library Quantification Kit. Store in the dark at −20°C. Do not freeze and thaw more than twice.
-
d.Run the following protocol on the qPCR machine: approx. 2.5 h
qPCR cycling conditions
Steps Temperature Time Cycles Initial activation/denaturation 95°C 5 min 1 Denaturation 95°C 30 s 35× Annealing/Extension/Data acquisition 60°C 45 s Optional: Run a melt curve analysis. -
e.Confirm that the reaction efficiency is between 90%–110% by analyzing the standard curve.
-
f.Calculate the library concentration as follows:
Library concentration as determined by qPCR
Library dilution Library concentration by qPCR in pM
Mean concentration in pM Dilution factor Rep1 Rep2 Rep3 1:500 x1 x2 x3 X d1 = 500 1:1000 y1 y2 y3 Y d2 = 1000 1:2000 z1 z2 z3 Z d3 = 2000 The average standard fragment length fStd (standards from the KAPA Library Quantification Kit) is 452 bp. The average size-selected library fragment length fLib (Pippin Prep) is 350 bp. The molecular weight of one base pair (Mbp) is approximated with 660 g/mol. The ChIP library volume (VLib) is 20 μl (see point 67e). -
g.Determine the number of amplification cycles required from the amount of ChIP DNA (ng) with the help of the following table from the KAPA Library Quantification Kit (Roche):
PCR amplification cycles
m(undiluted library in ng) Number of cycles <0.002 18 0.002–0.007 17 0.007–0.01 16–15 0.015–0.03 14 0.03–0.07 13 0.07–0.09 12 0.1–0.2 11 0.2–0.3 10 0.3–0.5 9 0.5–1 8 1–2 7 2–3 6 3–5 5
-
a.
CRITICAL: Take care to avoid too many rounds of amplification in order to prevent PCR duplicates from making up most of your sequencing reads. See Troubleshooting 6.
-
69.Library amplification
-
a.For each sample, prepare the following PCR reaction on ice:
Library amplification reaction
Reagent Amount Adapter-ligated library 20 μL 2× KAPA HiFi HotStart ReadyMix 25 μL 10× Library Amplification Mix 5 μL Total 50 μl All reagents are from the KAPA HyperPrep Kit. -
b.Run the following PCR program:
PCR cycling conditions
Steps Temperature Time Cycles Initial activation/denaturation 98°C 45 s 1 Denaturation 98°C 15 s X Annealing 60°C 30 s Extension 72°C 30 s Final extension 72°C 60 s 1 Hold 4°C Max 2 h
-
a.
Note: The number of cycles X has to be adapted according to the amplification cycles determined in 68g.
-
70.Post-amplification Clean-up with AMPure XP beads (1× AMPure XP bead clean-up)
-
a.Mix 50 μl of the amplified ChIP library with 50 μl of well mixed AMPure XP beads with a pipet by pipetting up and down for 10 times.
-
b.Proceed with the AMPure XP bead purification as described in 66b-g.
-
c.Add 16 μl Library Elution Buffer to the beads, remove the PCR tubes from the magnetic rack and pipet up and down for 10 times. Incubate for 2 min at 18°C–24°C.
-
d.Place PCR tubes into the magnetic tube holder and wait until the liquid is cleared (1–5 min).
-
e.Transfer 14 μl of the supernatant to a fresh PCR tube. This is the final library.
-
a.
Pause point: The final library can be stored at −20°C for up to 1 year. Take aliquots for Bioanalyzer and Qubit concentration measurement before freezing. Avoid freeze and thaw cycles.
-
71.
Perform quality control with the Agilent Bioanalyzer.
The Bioanalyzer profile on the left in Figure 5 shows an ideal library with a size distribution between 200 and 600 bp and no adapter or primer dimer contaminations:
Figure 5.
Profiles of libraries from H3K4me2 ChIP-seq in RAW264.7 cells on an Agilent Bioanalyzer 2010
Left: Setd1aDel/+ cells (Del). Right: Wild type cells (wt). The arrowhead points at contaminating adapters that need to be removed by another round of AMPure XP bead selection (see Troubleshooting 4).
In case of adapter contamination, see Troubleshooting 4.
-
72.Pooling of libraries for sequencing
-
a.Measure the library concentrations by Qubit.
-
b.Calculate the volume of each library to be pooled for sequencing as follows:
VLib – volume to pool for the individual library in μlNlanes – number of lanescpool – concentration of the final pool in μMVpool – volume of the final pool in μlMbp – molecular weight of one base pair (approx. 660 g/mol)fLib – average library size as determined by the BioanalyzerNsamples per lane – number of samples to pool per lane (see notes)cLib – concentration of individual library in ng/μl as measured by Qubit -
c.Fill to the Vpool with nuclease-free water and store the pool of ChIP libraries at −20°C until sequencing.Note: Flow cells and pooling strategy need to be adapted according to the number of libraries, the sequencer available and the desired sequencing depth. We recommend contacting the sequencing facility or provider before pooling. The number of samples to pool in one lane is determined by the total number of reads given by a flow cell (e.g., 800 M reads for one lane on the SP flow cell for the NovaSeq 6000) and the desired sequencing depth. Sequencing depth recommendations for the mouse genome are as follows:
Sequencing depth
ChIPed factor Desired sequencing depth Transcription factor 30–40 M Narrow histone modifications, RNA Polymerase II 60 M Broad histone modification s 80 M Input 200 M
-
a.
Expected outcomes
ChIP-seq libraries with 1–5 ng/μl size-selected DNA can be expected. If a library meets the following quality control standards, it can be processed for pooling and sequencing.
First, the ChIP-qPCR shows a significant enrichment at a positive locus when compared to a negative region, for both the target and the spike-in genome (see Figure 6).
Secondly, the bulk of the fragmented chromatin used for library preparation is between 200 and 1,500 bp in size. The size-selected library therefore resembles the majority of the fragmented chromatin.
Third, the fragment length distribution of the library resembles a shape similar to the library visualized in the Bioanalyzer profiles in Figure 5 left or broader. A homogeneous distribution of fragments from 200 to 600 bps is expected. No adapter dimers are detected.
Quantification and statistical analysis
Normalization of ChIP-qPCR results
Analyze ChIP-qPCR data by calculating the percentage input (%input) for each IP and for each locus (target and spike-in) as described in the following section. Here, we present an example of ChIP-qPCR against H3K4me2 in murine macrophages stimulated with LPS (L) or LPS plus Dexamethasone (L+D). 25% of spike-in chromatin from Drosophila S2 cells was used. In this example, the %input was calculated separately for each replicate. Fkbp5 represents the positive locus of the target genome and eRF3 denotes the positive locus of the spike-in genome. NegPol2 is a negative locus in the target genome.
-
1.
First, determine the performance of the primers used, by calculating the PCR efficiency from the standard curve.
Find the mean Ct values (technical replicates) of each standard in Table 1. Standards were prepared as described in step 50.
Table 1.
Ct values for standard (S) curve and non-template control (NTC). df – dilution factor
| Ct values | Fkbp5 | NegPol2 | eRF3 | ||
|---|---|---|---|---|---|
| log2(df) | Df | NTC | 36.21 | 33.56 | 36.19 |
| 13.29 | 10000 | S1 | 22.09 | 20.73 | 21.36 |
| 9.97 | 1000 | S2 | 25.23 | 23.81 | 24.68 |
| 6.64 | 100 | S3 | 28.55 | 26.98 | 28.11 |
| 3.32 | 10 | S4 | 33.05 | 29.71 | 32.76 |
| 0.00 | 1 | S5 | 34.06 | 33.24 | 38.34 |
| PCR efficiency in % | 94.0 | 91.2 | 106.7 | ||
We plot the standard curve as linear regression of the mean Ct values (technical replicates) over the log2-transformed dilution factor (Figure 6) and determine the PCR efficiency from the slope of the standard curve as follows.
The linear range of the standard curve is defined as the Ct value range, with the standard curve being linear. In our example, all the primers have a linear range from S1 (22 for Fkbp5) to S4 (33 for Fkbp5). S5 was excluded, as it was either outside the linear range or within one Ct value of the non-template control (Tab. 1).
-
2.
Adjust the input to 100% using the following formula:
inpadj – adjusted inputdf – dilution factor (500 for 1% input diluted 1:5 during qPCR) – mean Ct value of the qPCR replicates from the input sample
-
3.
Calculate the percentage input using the following formula:
– mean Ct value of the qPCR replicates for the ChIP sample
Table 2 shows the example data from the ChIP-qPCR against H3K4me2 in murine macrophages.
Table 2.
Spike in normalization for H3K4me2 in murine macrophages after LPS (L) or LPS+ Dexamethasone (L+D) treatment
| Raw Ct value |
%input |
|||||
|---|---|---|---|---|---|---|
| Fkbp5 | NegPol2 | eRF3 | Fkbp5 | NegPol2 | eRF3 | |
| IgG1 L+D_1 | 32.32 | 30.17 | 32.00 | 0.047 | 0.080 | 0.042 |
| IgG1 L+D_2 | 33.55 | 29.66 | 32.27 | 0.020 | 0.115 | 0.035 |
| IgG1 L+D_3 | 32.37 | 29.51 | 31.20 | 0.045 | 0.127 | 0.074 |
| IgG2 L+D_1 | 34.88 | 29.76 | 32.23 | 0.008 | 0.107 | 0.036 |
| IgG2 L+D_2 | 33.26 | 30.92 | 32.01 | 0.024 | 0.048 | 0.042 |
| IgG2 L+D_3 | 33.31 | 29.70 | 32.4 | 0.023 | 0.112 | 0.032 |
| H3K4me2_1 L+D_1 | 23.10 | 27.66 | 23.41 | 28.087 | 0.459 | 16.429 |
| H3K4me2_2 L+D_2 | 22.78 | 27.25 | 23.51 | 35.062 | 0.610 | 15.273 |
| H3K4me2_3 L+D_3 | 23.17 | 27.24 | 23.54 | 26.850 | 0.617 | 14.950 |
| H3K4me2_1 L+D_1 | 25.25 | 28.79 | 25.31 | 6.324 | 0.210 | 4.407 |
| H3K4me2_2 L+D_2 | 24.57 | 28.39 | 25.11 | 10.146 | 0.277 | 5.028 |
| H3K4me2_3 L+D_3 | 24.48 | 29.28 | 25.05 | 10.829 | 0.150 | 5.256 |
| Input L+D 50%_1 | 22.35 | 20.93 | 21.78 | |||
| Input L+D 50%_2 | 22.33 | 20.85 | 21.78 | |||
| Input L+D 50%_3 | 22.13 | 20.92 | 21.85 | |||
| IgG1 L_1 | 33.37 | 29.59 | 37.74 | 0.015 | 0.082 | 0.000 |
| IgG1 L_2 | 33.33 | 29.24 | 43.52 | 0.015 | 0.105 | 0.000 |
| IgG1 L_3 | 32.53 | 29.46 | 31.86 | 0.027 | 0.090 | 0.042 |
| IgG2 L_1 | 34.91 | 32.30 | 32.41 | 0.005 | 0.012 | 0.029 |
| IgG2 L_2 | 32.79 | 30.84 | 33.37 | 0.022 | 0.034 | 0.014 |
| IgG2 L_3 | 33.62 | 30.47 | 32.93 | 0.012 | 0.045 | 0.020 |
| H3K4me2_1 L_1 | 24.20 | 27.85 | 25.01 | 8.847 | 0.277 | 4.907 |
| H3K4me2_2 L_2 | 24.23 | 28.13 | 25.07 | 8.628 | 0.229 | 4.702 |
| H3K4me2_3 L_3 | 24.17 | 27.51 | 24.83 | 9.026 | 0.352 | 5.553 |
| H3K4me2_1 L_1 | 22.92 | 26.54 | 23.62 | 21.409 | 0.686 | 12.888 |
| H3K4me2_2 L_2 | 22.88 | 27.49 | 23.71 | 22.057 | 0.356 | 12.138 |
| H3K4me2_3 L_3 | 22.84 | 27.20 | 23.69 | 22.646 | 0.436 | 12.263 |
| Input L_1 50% | 21.64 | 20.22 | 21.34 | |||
| Input L_2 50% | 21.66 | 20.30 | 21.43 | |||
| Input L_3 50% | 21.79 | 20.56 | 22.22 | |||
| Mean of Input Samples | ||||||
| Fkbp5 | NegPol2 | eRF3 | ||||
| Input L+D | 22.270 | 20.897 | 21.801 | |||
| Input L | 21.698 | 20.359 | 21.664 | |||
| Adjusted Input to 100% | ||||||
| Fkbp5 | NegPol2 | eRF3 | ||||
| Input L+D | 21.270 | 19.897 | 20.801 | |||
| Input L | 20.698 | 19.359 | 20.664 | |||
Raw Ct values and %input.
-
4.
Continue to spike-in normalization using the following formula:
- %input of a locus occupied by the protein of interest in the target genome (positive)
- %input of a locus occupied by the protein of interest in the spike-in genome (positive)
Table 3 illustrates the example data for a H3K4me2 ChIP-qPCR experiment in murine macrophages.
Table 3.
Spike in normalization for H3K4me2 in murine macrophages after LPS (L) or LPS+ Dexamethasone (L+D) treatment
| Mean %input |
Spike-in Norm %inp |
||||
|---|---|---|---|---|---|
| Fkbp5 | NegPol2 | eRF3 | Fkbp5 | ||
| IgG1 L+D | 0.037 | 0.107 | 0.050 | H3K4me2_1 L+D_1 | 1.71 |
| IgG2 L+D | 0.018 | 0.089 | 0.036 | H3K4me2_2 L+D_2 | 2.30 |
| H3K4me2_1 L+D | 30.000 | 0.562 | 15.550 | H3K4me2_3 L+D_3 | 1.80 |
| H3K4me2_1 L+D | 9.099 | 0.212 | 4.897 | H3K4me2_1 L+D_1 | 1.43 |
| IgG1 L | 0.019 | 0.093 | 0.014 | H3K4me2_2 L+D_2 | 2.02 |
| IgG2 L | 0.013 | 0.030 | 0.021 | H3K4me2_3 L+D_3 | 2.06 |
| H3K4me2_1 L | 8.834 | 0.286 | 5.054 | H3K4me2_1 L_1 | 1.80 |
| H3K4me2_1 L | 22.037 | 0.493 | 12.430 | H3K4me2_2 L_2 | 1.83 |
| H3K4me2_3 L_3 | 1.63 | ||||
| SD %input | H3K4me2_1 L_1 | 1.66 | |||
| Fkbp5 | NegPol2 | eRF3 | H3K4me2_2 L_2 | 1.82 | |
| IgG1 L+D | 0.015 | 0.024 | 0.020 | H3K4me2_3 L_3 | 1.85 |
| IgG2 L+D | 0.009 | 0.035 | 0.004 | ||
| H3K4me2_1 L+D | 4.427 | 0.089 | 0.777 | Mean spike-in Norm | |
| H3K4me2_1 L+D | 2.427 | 0.063 | 0.439 | Fkbp5 | |
| IgG1 L | 0.006 | 0.011 | 0.024 | H3K4me2_1 L+D | 1.93 |
| IgG2 L | 0.008 | 0.016 | 0.007 | H3K4me2_1 L+D | 1.84 |
| H3K4me2_1 L | 0.199 | 0.062 | 0.444 | H3K4me2_1 L | 1.75 |
| H3K4me2_1 L | 0.618 | 0.172 | 0.402 | H3K4me2_1 L | 1.77 |
| SD Spike-in Norm | |||||
| Fkbp5 | |||||
| H3K4me2_1 L+D | 0.32 | ||||
| H3K4me2_1 L+D | 0.35 | ||||
| H3K4me2_1 L | 0.11 | ||||
| H3K4me2_1 L | 0.10 | ||||
SD - standard deviation, Norm-normalized.
Figure 7 shows the enrichment of H3K4me2 in LPS (L) and LPS plus Dexamethasone (L+D) treated macrophages at a positive (Fkbp5) and a negative locus (Negpol2) before (Figure 7A) and after spike-in normalization (Figure 7B). Before spike-in normalization, the results from both replicates are highly variable, both in LPS and LPS plus Dexamethasone stimulated macrophages. A similar variation is observed for the Drosophila spike-in locus eRF3 pointing towards a technical bias between the different test tubes (Figure 7A). By normalization to the positive spike-in locus (eRF3), we are able to account for this variation (Figure 7B).
CRITICAL: The spike-in ratio must be sufficiently high to measure reliable Ct values by qPCR (see Troubleshooting 5).
Figure 7.
Spike-in ChIP-qPCR of H3K4me2 in murine macrophages stimulated with LPS (L) and LPS plus Dexamethasone (L+D)
(A) Non-normalized ChIP-qPCR results showing the percentage input for the Fkbp5 (positive) and NegPol2 (negative) loci in the murine genome and the percentage input for the eRF3 locus (positive for the spike-in genome). A ChIP against IgG is included as additional negative control.
(B) Spike-in normalized enrichment of the Fkbp5 locus. The experiment was performed in duplicates. Error bars represent the standard deviation of the qPCR triplicates.
Normalization of ChIP-seq results
The quality of the sequencing reads can be evaluated by FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Samples not yielding at least 50% of the required sequencing depth (see 72c) should be submitted for re-sequencing. The “per base quality score” should be above 28 for all read positions. Low quality reads are filtered out during sample processing with Samtools (Li et al., 2009). Adapter sequences are removed with Trimmomatic (Bolger et al., 2014).
Samples are processed via a standard ChIP-seq pipeline (Figure 8 left). Each sample is mapped against the target genome (Mus musculus Ensembl genome build GRCm38.p6 (mm10)) (Cunningham et al., 2019) and against the spike-in genome (Drosophila melanogaster Ensembl BDGP6 release 78 (dm6)) in this example (Cunningham et al., 2019)) using BWA-MEM (Li, 2013). Peaks are called using MACS2 (Zhang et al., 2008). For more details, see Mir et al. (Mir et al., 2019) and the ENCODE pipeline (https://www.encodeproject.org/data-standards/chip-seq/ (Landt et al., 2012)).
Figure 8.
ChIP-seq pipeline and scaling by spike-in chromatin for differential occupancy analysis
Left: Standard ChIP-seq pipeline, run separately against the mm10 (target) and the dm6 (spike-in) reference genomes. Filters are indicated in red, tasks in blue. Right: Spike-in normalization. RQC – number of quality-filtered reads; RiP – number of reads overlapping peaks; IPeff – IP efficiency; dm – Drosphila melanogaster (spike-in genome); mm – Mus musculus (target genome); sf – scale factor; abs – absolute; rel – relative, max - maximum
Samples with fewer than 80% of reads mapping to the target genome or more than 80% of PCR duplicates (see Troubleshooting 6) should be excluded from analysis. Furthermore, ChIP-seq samples of transcription factors with <1% of reads overlapping peaks (RiP), or histone marks with <10% RiP in either target genome or spike-in genome should be removed as well (see Troubleshooting 7). Samples with a recovery of <1% spike-in DNA (of all non-duplicated and aligned reads) were removed, too. Note that the proportion of recovered spike-in reads will depend on the target and the spike-in genome size ratio and the genomic coverage of the protein of interest (see Troubleshooting 5).
The peak union of all replicates (peak universe) can be generated in R (https://www.R-project.org/
(Team, 2017)) using the GenomicRanges package (Lawrence et al., 2013), and blacklisted regions can be removed using the following script:
Note: At the time of publications, BED files of the updated blacklisted regions could be found at https://github.com/Boyle-Lab/Blacklist/tree/master/lists.
The number of reads covering peaks (RiP) is determined using BEDTools (Quinlan and Hall, 2010)
and the R script below:
The immunoprecipitation efficiency (IPeff) is calculated from the reads covering peaks (RiP) divided by all quality-filtered reads (RQC) for each reference genome. The absolute scale factor (sfabs) for each sample is calculated from the percentage of spike-in chromatin (100∗spike-in reads (Rdm)/(spike-in reads (Rdm) + target reads (Rmm))) divided by the RiP for the spike-in genome and normalized by the IP efficiency ratio between spike-in and target genome (IPeffdm/IPeffmm) (see Figure 8 right).
The absolute scale factors of all samples are set into relation (sfrel) by normalizing to the highest occurring absolute scale factor among all samples. Those relative scale factors are used for scaling BigWig files with deepTools (Ramirez et al., 2014):
The inverse of the relative scale factors replaces the sizeFactors in DESeq2 (Love et al., 2014) for differential occupancy analysis in R.
Alternatives: Diffbind (Ross-Innes et al., 2012) also offers spike-in normalization and differential occupancy analyses for simple designs.
Note: DESeq2 allows the usage of complex experimental designs.
Table 4 includes the mapping statistics (%map), the calculated IP efficiencies (IPeff), the absolute (abs.) and the relative (rel.) scale factors (sf) of an example data set (Greulich et al., 2021). FASTQ files are deposited on GEO with the accession number GEO: GSE138017.
Table 4.
Scale factor calculations and mapping statistics for H3K4me2 in wild type (wt) and Setd1aDel/+ (Del) RAW264.7 cells after LPS (L) or LPS plus Dexamethasone (L+D) treatment
| Sample ID | Geno | treat | #Reads | %map mm10 | %map dm6 | RQC mm10 | %Dupl. mm10 | RQC dm6 | %Dupl. dm6 | RiP mm10 | IPeff mm10 | RiP dm6 | IPeff dm6 | Abs. Sf | Rel. Sf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSM4096676 | Wt | LPS | 150594152 | 98.0 | 1.1 | 55647040 | 62.3 | 750324 | 53.1 | 9015482 | 0.16 | 217412 | 0.29 | 10.94 | 1.000 |
| GSM4096677 | Wt | L+D | 273648766 | 98.3 | 1.5 | 132706914 | 51.1 | 1979352 | 52.3 | 41604331 | 0.31 | 610549 | 0.31 | 2.37 | 0.216 |
| GSM4096678 | Wt | L+D | 178799812 | 98.3 | 2.0 | 106874214 | 39.2 | 2281740 | 41.6 | 31827833 | 0.30 | 552688 | 0.24 | 3.08 | 0.281 |
| GSM4096679 | Del | LPS | 203958426 | 98.3 | 2.4 | 156348392 | 22.1 | 3315942 | 30.8 | 19587761 | 0.13 | 629492 | 0.19 | 5.00 | 0.457 |
| GSM4096680 | Del | LPS | 210208094 | 98.2 | 1.7 | 75365880 | 63.5 | 1758958 | 52.2 | 18737752 | 0.25 | 482824 | 0.27 | 5.22 | 0.477 |
| GSM4096681 | Del | L+D | 243718250 | 98.3 | 1.7 | 69405402 | 71.0 | 1826934 | 55.8 | 23061949 | 0.33 | 509125 | 0.28 | 4.22 | 0.386 |
| GSM4096683 | Wt | L+D | 114420802 | 95.8 | 3.2 | 46958500 | 57.2 | 1949396 | 46.6 | 12314423 | 0.26 | 550129 | 0.28 | 7.80 | 0.712 |
| GSM4096684 | Wt | LPS | 113331148 | 96.7 | 2.6 | 45138652 | 59.5 | 1388290 | 53.2 | 13706729 | 0.30 | 469364 | 0.34 | 7.08 | 0.647 |
| GSM4096685 | Del | L+D | 77635734 | 96.4 | 3.6 | 36757906 | 50.9 | 1207134 | 56.8 | 12829586 | 0.35 | 539918 | 0.45 | 7.55 | 0.690 |
| GSM4096686 | Del | LPS | 143159666 | 94.8 | 3.2 | 64833572 | 52.3 | 2469554 | 45.9 | 23516324 | 0.36 | 1077635 | 0.44 | 4.10 | 0.374 |
treat – treatment; geno – genotype, Dupl. - duplicates
The effect of spike-in normalization on the read counts across all H3K4me2 peaks (peak universe) between replicates, and examples of normalized versus non-normalized genome browser tracks, are shown in Figure 9.
Figure 9.
Spike-in normalization of ChIP-seq data
(A) Tag counts per peak (resized to 1 kb) for the H3K4me2 peak union from two biological replicates of H3K4me2 ChIP-seq in LPS plus Dexamethasone treated wild type (left, gray) and Setd1aDel/+ (Del, right, red) RAW264.7 cells. The upper plots show raw tag counts, whereas the lower plots show the tag counts after spike-in normalization. Dashed lines indicate the linear regression (gray) and ideal regression lines (red), if both samples were identical. RS – Spearman correlation coefficient. p – Significance of the correlation. Rep – replicate.
(B) Example genome browser tracks of the Dusp1 (up) and Tsc22d3 (down) loci for the same samples as in A. Visualization with Integrative Genome Browser (Freese et al., 2016). Gray shadows indicate peaks with reduced inter-replicate variation after spike-in normalization. Colors as in A.
Limitations
ChIP depends on the specificity and availability of the antibody. Any antibody used in ChIP experiments needs to be extensively tested by ChIP-qPCR in the cell type or tissue of interest, ideally by comparing wild type and knockout cells. A pre-screen by Western blot is possible. If knockout cells/tissues are not available, specific blocking peptides might be purchased and included as controls. The latter is recommended when profiling closely related histone modifications to ensure that the ChIP antibodies do not cross-react. Additionally, ChIP experiments require a certain amount of starting material. For low amounts of input material, Cut&Run or Cut&Tag techniques are an alternative, but come with their own limitations (Kaya-Okur et al., 2019; Skene and Henikoff, 2017). ChIP-seq experiments in particular are limited to the availability of genome sequences for the target species.
Heterologous spike-in ChIP experiments have several additional technical limitations. For heterologous spike-in ChIP, the protein of interest needs to be conserved between the target and the spike-in species. Closely related species can be used for ChIP-qPCR, given that the primers were tested for cross-reactivity, but are not recommended for ChIP-seq experiments. To test for cross-mappability of the spike-in species, obtain the reference genome for the spike-in species, bin it into fragments matching the sequencing read length (e.g., 50 or 101 bp) and map it against the target species, and vice versa. Less than 10% cross-mappability are acceptable for heterologous spike-in experiments. On top of the target protein conservation, the antibody of interest needs to react with the protein epitope of interest across species. The reactivity of the ChIP antibody needs to be tested in cross-linked tissue or cell material form the target species as well as for the spike-in cells or tissues. For this purpose, one can design primers against positive and negative regions in both genomes and test several antibodies against the protein of interest for specificity by ChIP-qPCR.
Note: Polyclonal antibodies are more likely to react with the protein of interest across species.
Troubleshooting
Problem 1
Target and spike-in cells have different sonication conditions (step 6).
Potential solution
Sonication conditions have to be optimal for each of the tissues/cells used as target or spike-in. Therefore, optimal sonication conditions need to be established before the start of the experiment. In the case of murine macrophages and Drosophila S2 cells, sonication conditions are very similar (12 cycles for macrophages and 10 cycles for S2 cells at high settings, see Figure 1). Thus, both cell types can be mixed at the desired spike-in ratio before sonication. This might not always be the case.
If the sonication conditions differ substantially between target and spike-in species, lyse and shear both cell types separately with the optimal conditions for each sample. Mix the fragmented chromatin at the desired spike-in ratio, after removing insoluble chromatin (step 18).
Problem 2
Find a ChIP-grade antibody (Before you begin, step 22).
Potential solution
A key success factor for ChIP experiments is a specific antibody. Find below some recommendations on how to select a good ChIP antibody.
-
•
Acquire ChIP-validated antibodies, but still confirm the specificity of those antibodies by performing ChIP-qPCR against a positive and negative locus in the specific experimental system (e.g., cell type or tissue) as described in the “Before you begin” section.
-
•
Try antibodies suitable for immunohistochemistry or immunofluorescence of formaldehyde-fixed tissues.
-
•
Try polyclonal antibodies, as they may have a higher probability of recognizing an unmasked epitope and as they may exhibit cross-species specificity.
-
•
Test each antibody against a knockout or knockdown cell line if available, or use blocking peptides to confirm antibody specificity.
-
•
Include more than one negative (not bound by the protein of interest) and positive locus, as well as an IgG control, to test for specificity in Chip-qPCR.
Problem 3
Undetectable ChIP DNA (step 62).
Potential solution
The sensitivity of the Qubit dsDNA HS kit lies between 0.2–100 ng of DNA. A ChIP sample might be undetectable by Qubit.
-
•
Repeat the ChIP and combine the eluted DNA samples from two ChIP experiments.
-
•
If the input material is limiting a repetition of the experiment, process the complete ChIP DNA for library preparation. After library quantification, do not add more than 14 PCR cycles to avoid over amplification of the library. A specific enrichment for the factor of interest must have been observed by ChIP-qPCR (see Figure 7), and the majority of the chromatin fragments must lie between 150 and 1500 bp, as determined by agarose gel electrophoresis or Bioanalyzer of the input DNA (see Figures 1 and 3).
Problem 4
Adapter contamination of the final library.
Potential solution
The Bioanalyzer profiles might reveal an additional peak at approx. 120 bp similar to Figure 5. Those are adapter dimers contaminating the library. In contrast to the remaining PCR primers at around 35 bp, adapter dimers need to be removed before sequencing, as they contain the adapter sequence able to bind to the flow cell.
Repeat post amplification clean-up step 70.
Note that any additional clean-up step will result in the loss of approximately half the concentration of the final library.
Problem 5
Low amount of spike-in DNA after ChIP (step 6).
Potential solution
If the spike-in qPCR yields very high Ct values or N/As in the ChIP samples, or if they fall below 1% of all non-duplicated and properly aligned reads for ChIP-Seq samples, the concentration of the spike-in chromatin is too low to allow for proper estimation of technical bias.
-
•
Make sure the spike-in primers are optimal, by determining at the PCR efficiency and the melt curve. The melt curve should only show one prominent peak. The PCR efficiency should lie between 90 - 100% for a Ct value range, recapitulating the Ct values of the ChIP samples.
-
•
If the spike-in primers are optimized and show significant enrichment for the input sample, the chosen positive locus might not be positive.
-
•
If the input sample also shows a high Ct value for the spike-in chromatin, or if < 1% spike-in reads are detected in ChIP-Seq, consider increasing the spike-in ratio up to 25% in ChIP-qPCR or up to 10% in ChiP-Seq experiments.
Problem 6
High number of PCR duplicates (step 69).
Potential solution
PCR duplication rates >50% are not only expensive, but they also bias the occupied chromatin fraction towards smaller fragments. A true PCR duplicate has the same start and end of the sequencing read. In order to differentiate PCR duplicates from naturally occurring duplicates, paired-end sequencing is recommended. PCR duplicates mostly arise from library amplification (step 69), when more than 7 amplification cycles were performed.
-
•
Limit PCR cycles to maximum 7 cycles.
-
•
If sufficient ChIP DNA is available, consider combining two library preparations of the same sample, with low numbers of amplification cycles each.
-
•
If sufficient ChIP DNA was obtained, insufficient adapter annealing might cause a requirement for extra PCR cycles during library amplification. Amplification primers only bind adapters and therefore, only adapter-ligated chromatin fragments will be amplified. To ensure optimal adapter ligation, avoid freeze and thaw cycles of adapter dilutions and make sure that the T4 ligase is stored properly. If the problem persists, reorder adapter stocks or prepare fresh adapter dilutions.
Problem 7
High background signal (steps 5, 7, 22, and 26).
Potential solution
The fraction of reads mapping into peaks reflects the efficiency of the ChIP (IP efficiency) and serves as quality readout that reports on how many reads were specifically purified. IP efficiencies below 10% indicate an unspecific recovery of chromatin fragments, due to various reasons. Similarly, a low enrichment for a positive locus over the negative locus in ChIP-qPCR might indicate unspecific binding, even if the IgG control was not affected.
There are several reasons for a high background signal.
If the antibody causes an unspecific signal (see Troubleshooting 2), the enrichment for a positive locus over the negative locus (region not bound by the protein of interest) will be low, and both signals will be significantly higher than the signal observed in the IgG control.
Besides an unspecific antibody, the antibody-epitope interaction may be inhibited by the SDS content of the shearing buffer. Try a low-SDS shearing buffer (step 5) and add Triton X-100 up to 1%, to sequester the SDS during immunoprecipitation (step 21).
| Low-SDS shearing buffer | Final concentration | Amount |
|---|---|---|
| SDS (20%)∗ | 0.1% vol/vol | 5 mL |
| Sodium deoxycholate (10%)∗ | 0.1% vol/vol | 10 mL |
| EDTA pH=8.0 (0.5 M) | 1 mM | 2 mL |
| Tris-HCl pH=8.0 (1 M)∗ | 50 mM | 50 mL |
| Triton X-100 | 1% vol/vol | 10 mL |
| ddH2O | N/A | 880 mL |
| Total | 1 l |
Low-SDS Shearing Buffer is stable at 18°C−24°C for up to 6 month.
∗Sodium deoxycholate is harmful when swallowed and may cause respiratory irritation. Avoid breathing dust/fume/vapors or sprays. Do not eat/drink or smoke while using Sodium Deoxycholate. Wash hands after handling. Wear protective clothing. Work in a well-ventilated area. Dispose according to regional/national regulations.
Note: When changing the shearing buffer, sonication conditions have to be re-established.
“Over-sheared” chromatin displays unspecific binding to the beads or interactions of the ChIP antibody with alternative epitopes. Confirm fragment sizes after sonication, and do not proceed with ChIP if most of the chromatin fragments are below 500 bp after fast-reverse cross-link.
Unspecific binding of chromatin and DNA to the beads may cause increased background signal. Try to reduce the incubation time with the beads. Otherwise, pre-clearing of chromatin with pre-blocked Dynabeads or sepharose beads might remove proteins and DNA that binds unspecifically to the beads. Add 20 μl of pre-blocked beads to each sample and incubate for 1 h at 4°C while rotating slowly. After 1 h, place tubes in a magnetic rack (Dynabeads) or spin for 30 s at 300 × g at 4°C (sepharose beads). Transfer the supernatant to a fresh tube and proceed with the immunoprecipitation (step 22).
Rarely, batch effects in the Dynabeads or sepharose beads might cause higher background signal and are resolved after purchase of a new batch.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Franziska Greulich (franziska.greulich@tum.de).
Material availability
This study did not generate any unique reagents. All reagents used are commercially available and listed in the Key Resource table.
Data and code availability
The example data used in this study is available from GEO (https://www.ncbi.nlm.nih.gov/geo/ (Edgar et al., 2002)) under the accession number GEO: GSE138017. All unique code is reported in this study.
Acknowledgments
We sincerely thank E. Graf, S. Loesecke, T. Schwarzmayr, and I. de la Rosa (HMGU genomics core) for their contribution to the NGS studies. We are grateful to I. Guderian and S. Regn for administrative assistance. This work received funding from the DFG (UH 275/1-1, SFB 1064 Chromatin Dynamics and TRR205 Adrenal Research to NHU and Entzuendungsprozesse GR 5179/1-1 to FG) and from the ERC (ERC-2014-StG 638573 SILENCE to NHU). Figures were partially created using BioRender.
Author contributions
F.G. designed, performed, and analyzed the NGS experiments. F.G. and A.M. developed, validated, and optimized the spike-in ChIP protocol. A.M. performed the spike-in ChIP-qPCR. T.H. optimized the library preparation protocol. F.G. and N.H.U. supervised the work and secured funding. A.M. and F.G. wrote the manuscript together with N.H.U. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
References
- Allhoff M., Sere K., J F.P., Zenke M., I G.C. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 2016;44:e153. doi: 10.1093/nar/gkw680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B., Yuan C.C., Craske M.L., Labhart P., Guler G.D., Arnott D., Maile T.M., Busby J., Henry C., Kelly T.K. An alternative approach to ChIP-seq normalization enables detection of genome-wide changes in histone H3 lysine 27 trimethylation upon EZH2 inhibition. PLoS One. 2016;11:e0166438. doi: 10.1371/journal.pone.0166438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese N.H., Norris D.C., Loraine A.E. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016;32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C.S.F. Avoiding adsorption of dna to polypropylene tubes and denaturation of short dna fragments. Technical Tips Online. 1998 [Google Scholar]
- Greulich F., Wierer M., Mechtidou A., Gonzalez-Garcia O., Uhlenhaut N.H. The glucocorticoid receptor recruits the COMPASS complex to regulate inflammatory transcription at macrophage enhancers. Cell Rep. 2021;34:108742. doi: 10.1016/j.celrep.2021.108742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A.A., Dyar K.A., Greulich F., Quagliarini F., Jouffe C., Hubert M.J., Hemmer M.C., Uhlenhaut N.H. In vivo ChIP-seq of nuclear receptors: a rough guide to transform frozen tissues into high-confidence genome-wide binding profiles. Methods Mol Biol. 2019;1966:39–70. doi: 10.1007/978-1-4939-9195-2_5. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P.J., Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Ye Z., Lenz S.W., Clark C.R., Bharucha A., Farrugia G., Robertson K.D., Zhang Z., Ordog T., Lee J.H. Purification of nanogram-range immunoprecipitated DNA in ChIP-seq application. BMC Genomics. 2017;18:985. doi: 10.1186/s12864-017-4371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The example data used in this study is available from GEO (https://www.ncbi.nlm.nih.gov/geo/ (Edgar et al., 2002)) under the accession number GEO: GSE138017. All unique code is reported in this study.