Summary
The interplay between the phytohormone abscisic acid (ABA) and the gasotransmitter nitric oxide (NO) regulates seed germination and post-germinative seedling growth. We show that GAP1 (germination in ABA and cPTIO 1) encodes the transcription factor ANAC089 with a critical membrane-bound domain and extranuclear localization. ANAC089 mutants lacking the membrane-tethered domain display insensitivity to ABA, salt, and osmotic and cold stresses, revealing a repressor function. Whole-genome transcriptional profiling and DNA-binding specificity reveals that ANAC089 regulates ABA- and redox-related genes. ANAC089 truncated mutants exhibit higher NO and lower ROS and ABA endogenous levels, alongside an altered thiol and disulfide homeostasis. Consistently, translocation of ANAC089 to the nucleus is directed by changes in cellular redox status after treatments with NO scavengers and redox-related compounds. Our results reveal ANAC089 to be a master regulator modulating redox homeostasis and NO levels, able to repress ABA synthesis and signaling during Arabidopsis seed germination and abiotic stress.
Keywords: abiotic stress, abscisic acid, redox homeostasis, nitric oxide, seed, transcription factor
Graphical abstract
Highlights
-
•
GAP1 encodes transcription factor ANAC089, expressed in seeds and stress
-
•
Changes in cellular redox status induce translocation of ANAC089 to the nucleus
-
•
ANAC089 binds specifically to genes controlling seed germination and abiotic stress
-
•
ANAC089 regulates NO levels and cell redox and represses ABA synthesis and signaling
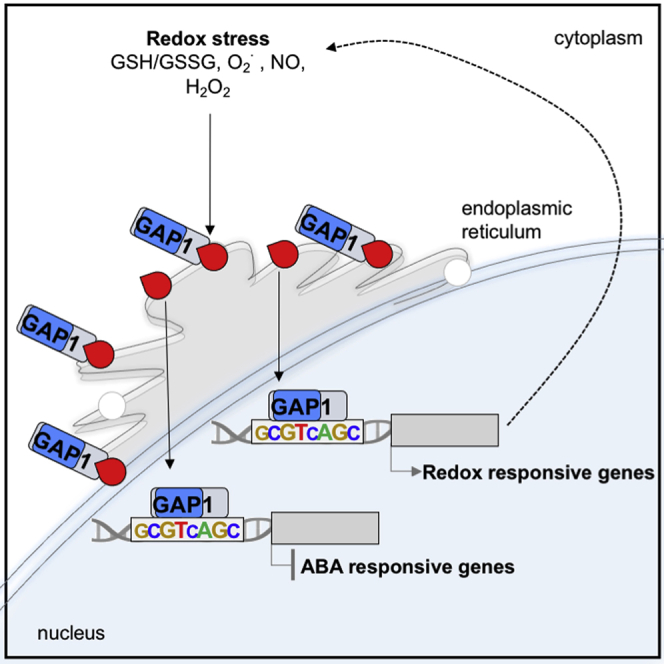
Albertos et al. report the cleavage of ANAC089 transcription factor from the membrane-anchoring domain in the endoplasmic reticulum, controlled by cell redox alterations. Once in the nucleus, ANAC089 induces redox homeostasis-related genes and represses ABA-responsive genes to increase NO levels and ABA insensitivity during seed germination and abiotic stress.
Introduction
Seed germination starts with water uptake by the dry seed, involving the reactivation of metabolic and gene expression events and finishes with the protrusion of the embryonic axis through the seed coat (Bewley and Black, 1994). This process is arguably one of the most important and vulnerable of a plant life cycle. The phytohormone abscisic acid (ABA) and the gasotransmitter nitric oxide (NO) are involved in regulating seed formation and germination (Bethke et al., 2004b; Finkelstein, 2013; Sanz et al., 2015).
ABA acts through a complex signaling cascade of transcription factors (TFs) to fine-tune ABA responses and induce changes in gene expression; thus, multiple stress responses are modulated by this pathway during seed germination (Nakashima and Yamaguchi-Shinozaki, 2013). Different ABA-inducible TFs include members of AREB/ABI5 (ABA-response element [ABRE]-binding proteins), VP1/B3/ABI3, AP2/ABI4, MYC, MYB, HD-ZIP, and NAC (NAM-ATAF1,2-CUC2) family proteins (Fujita et al., 2011). The NAC domain-containing proteins constitute a large family of plant-specific TFs that is represented by at least 117 members in Arabidopsis thaliana (Nuruzzaman et al., 2010). Interestingly, several Arabidopsis NACs, such as ATAF1, ATAF2, ANAC019, ANAC055, ANAC072/RD26, NAC2, and NTL8, are involved in ABA-dependent responses to abiotic stresses (drought, salt, cold) during different developmental windows (Fujita et al., 2004; He et al., 2005; Jensen et al., 2010; Jiang et al., 2009; Kim et al., 2008; Mao et al., 2012; Tran et al., 2004). Similarly, anac092-1, ntl8-1, and ntm2-1 mutants are positive regulators of seed germination under high salt conditions (Balazadeh et al., 2010; Kim et al., 2008; Park et al., 2011).
The crosstalk between NO and ABA signaling pathways during seed germination has been demonstrated by two different molecular mechanisms (Albertos et al., 2015; Gibbs et al., 2014; Sánchez-Vicente et al., 2019). On the one hand, NO promotes group ERF VII TFs degradation by the N-degron pathway, preventing ABI5 expression and enhancing seed germination (Gibbs et al., 2014). On the other hand, ABI5 S-nitrosation, a NO-dependent post-translational modification, triggers degradation of this central ABA repressor to promote seed germination and post-germination (Albertos et al., 2015). Additional control of ABA perception and signaling pathways by NO occurs at the level of PYR/PYL/RCAR receptors, which are inhibited by Tyr nitration (Castillo et al., 2015), enabling the activation of PP2C, which in turn inactivates SnRKs. Furthermore, the inhibition of the activity of SnRKs 2.2, 2.3, and 2.6 by S-nitrosation and SnRK2.6 downregulation by NO treatment, impairs seed germination and stomatal closure (Wang et al., 2015a, 2015b; Zhao et al., 2016). However, signaling components that regulate NO homeostasis in connection with ABA-dependent targets are currently unknown. Here, we report on the identification and molecular characterization of the redox-sensitive TF ANAC089 during seed germination and seedling establishment. We show that ANAC089 encodes an extranuclear localized NAC-domain TF with a crucial transmembrane domain (TM), which functions as an ABA synthesis and response repressor, regulates NO levels, and modulates cell redox-related homeostasis genes in seeds. The isolation of two anac089 gain-of-function mutants demonstrates the functional relevance of ANAC089 in the ABA and NO/redox transcriptional networks during seed germination and in stress responses. Moreover, we determine the DNA binding specificity of ANAC089 following a microarray-based approach. Information on ANAC089 binding specificity, together with bioinformatics analysis of the transcriptome data, associates this TF with target genes relevant to the germination and stress responses. We conclude that ANAC089 is a master regulator of seed germination and seedling development under abiotic stress.
Results
GAP1 encodes the ANAC089 TF expressed in dry seeds
To gain further insight into the regulation of seed germination by ABA and NO, we implemented genetic screening to identify gap (germination in ABA and 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide [cPTIO]) mutants (Albertos et al., 2015). Apart from 5 abi5 alleles, 2 other mutants, gap1-1 and gap1-2, were isolated from ethyl methane sulfonate (EMS)-mutagenized Landsberg erecta (Ler) and fast neutron-mutagenized Col-0 M2 seeds, respectively (Figure 1).
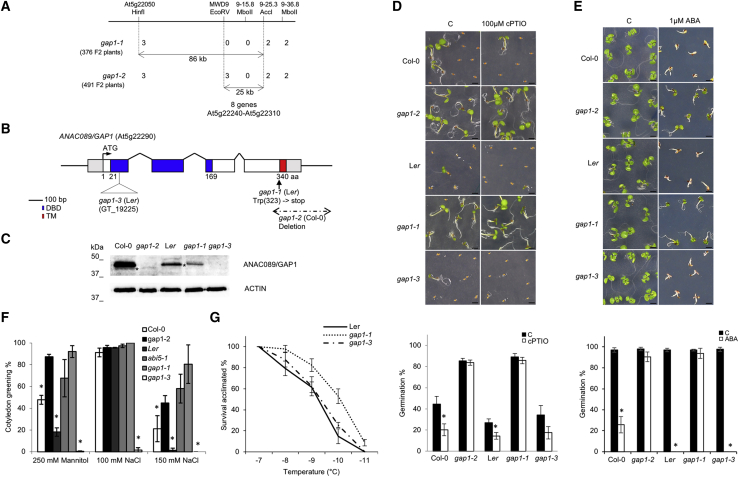
Figure 1.
Molecular characterization of gap1 encoding ANAC089 transcription factor during seed germination and stress
(A) Mapping based on Col/Ler polymorphisms in top arm of chromosome 5 where gap1-1 and gap1-2 mutation were located.
(B) Schematic representation of ANAC089 with the position of the 3 alleles gap1-1, gap1-2, and gap1-3. NAC domain (blue) and transmembrane domain (TM, red) are indicated.
(C) ANAC089 protein levels in 3-day-old imbibed seeds from indicated backgrounds. Asterisks indicate bands corresponding to ANAC089ΔC-2 and ANAC089ΔC-1 protein detected in gap1-2 and gap1-1 mutants. Actin protein levels are shown as a loading control.
(D) Insensitivity of gap1-1 and gap1-2 mutants to NO scavenging by cPTIO compared to the wild-type (Col-0, Ler) and gap1-3 mutant during seed germination and seedling establishment. Seeds were freshly harvested and sown in control media (C) or contain 100 μM cPTIO. Images were taken after 7 days and germination percentages 3 days after sowing. Scale bars, 1 mm.
(E) Insensitivity of gap1-1 and gap1-2 mutants to ABA compared to the wild-type (Col-0, Ler) and gap1-3 mutant during seed germination and seedling establishment. Seeds of the indicated genotypes were stratified for 3 days and sown in control media (C) or contain 1 μM ABA. Images were taken after 10 days and germination percentages 3 days after sowing. Scale bars, 1 mm.
(F) Stress germination assays in MS medium supplemented with either 250 mM mannitol or 100 and 150 mM NaCl.
(G) Freezing tolerance of 2-week-old cold-acclimated (7 d, 4°C) plants exposed to the indicated freezing temperatures for 6 h. Survival percentages were evaluated after 1 week of recovery at 22°C under long-day conditions.
In all of the graphs, each value represents the average germination percentage of 50–100 seeds, with error bars being the SEs of 3 replicates. Asterisks indicate significant differences compared with control versus treatment or Col-0 and Ler, respectively (t test, p < 0.05).
Both mutant lines were independently mapped, based on Col/Ler polymorphisms, to the top arm of chromosome 5. The gap1-1 mutation is located in an 86-kb region between cleaved amplified polymorphic sequence (CAPS) markers, At5g22050/HinfI and MWD9-25.3/AccI, and the gap1-2 mutation in a 25-kb region between MWD9/EcoRV and MWD9-25.3/AccI (Figures 1A and S1A). This shared 25-kb region contains 8 genes, and sequencing of them revealed that both mutants contained a mutation in the At5g22290. This gene encodes ANAC089, a NAC TF with a deduced protein of 340 amino acids (Molecular Mass ∼38 kDa), holding a TM on the C terminus (Figure 1B) (Ooka et al., 2003). The gap1-1 mutation substitutes Trp323 to a premature stop codon. Southern blot and PCR analysis suggested that the gap1-2 mutation was an ∼700-bp deletion that includes the C-terminal region (Figures S1A and S1B). In addition, the gap1-3 knockout ANAC089 mutant was identified as a gene trap insertional line (Figure S1C).
Our genetic studies indicate that mutations in the ANAC089 sequence in gap1-1 and gap1-2 alleles compromise the C-terminal part of the protein, generating the truncated versions lacking the TM ANAC089ΔC-1 and ANAC089ΔC-2, respectively (Figures 1B, 1C, and S1D). To test this fact, total protein extracted from the Arabidopsis Col-0, Ler and ANAC089 gain- and loss-of-function mutants was analyzed by immunoblotting using the anti-ANAC089 polyclonal antibody (Figure 1C). Genetic crosses between parental lines (Col-0) and the mutant (gap1-2) resulted in an ABA-insensitive phenotype of F1 progeny and point to gap1-2 as a dominant mutation (Figure S1E).
ANAC089 expression appears during seed maturation, rising up to the highest levels in dry seeds (Figure S1F). Quantitative RT-PCR was performed to examine the mRNA abundance of ANAC089 found to be accumulated in dry seed and reduced after 24 and 48 h of seed imbibition (Figures S1G and S1H), while 48-h ABA treatment induced the expression of ANAC089 in seeds (Figure S1I). Previous microarray analysis indicated that mRNA of the ANAC089 is the 7th most abundant in 740 mRNA species for TFs in Col-0 dry seeds (Preston et al., 2009). The gap1-2 mutant reduced transcript abundance of ANAC089 in both dry and imbibed seeds (Figure S1G). These ANAC089 expression patterns in seeds may be indicative of a variety of functions during this developmental stage.
gap1-1 and gap1-2 are gain-of-function mutants
Germination assays were performed to corroborate the ABA and cPTIO insensitivity of gap1-1 and gap1-2 alleles (Figures S2A–S2D). A dose-response assay with the NO scavenger cPTIO revealed that 100 μM was a suitable concentration to evaluate the maintenance of seed dormancy for further analysis (Figures S2A and S2B). Freshly harvested seeds of Col-0, Ler, and gap1 mutants were collected and directly sown on mannitol salt (MS) agar plates untreated (control) or supplemented with 100 μM cPTIO. gap1-1 and gap1-2 mutants were not dormant and displayed insensitive phenotypes to endogenous NO depletion by cPTIO, which prevented seed germination in wild-type and gap1-3 knockout mutants (Figure 1D). The effect of different ABA concentrations on the inhibition of seed germination was also analyzed with significant differences at 0.5 and 1 μM ABA (Figures S2C and S2D). Thus, 3-day-old stratified seeds from Col-0, Ler, and gap1 mutants were sown in media supplemented with 1 μM ABA. As expected from the screening, gap1-1 and gap1-2 mutants exhibited ABA-insensitive phenotypes during seed germination and seedling establishment in contrast to the inhibitory response to ABA in wild-type and gap1-3 knockout mutant (Figure 1E). To delve into the role of ANAC089 and NO homeostasis during seed germination, combinations of ABA together with cPTIO or S-nitrosoglutathione (GSNO) were analyzed in the wild-type (Col-0, Ler) and gap1 mutants. The dominant mutants gap1-1 and gap1-2 still showed a clear insensitive phenotype to a combined treatment of ABA plus cPTIO (Figures S2E and S2G). Regarding the combination of ABA plus GSNO, the cotyledon greening was greatly improved in the wild-type Col-0, showing that GSNO is able to counteract the action of ABA, as previously reported (Albertos et al., 2015). For gap1-1 and gap1-2 mutants, the cotyledon greening reached 100%, while Ler and gap1-3 showed higher ABA sensitivity, and the GSNO treatment was unable to promote seed germination and seedling development (Figures S2F and S2G). The fact that gap1-1 and gap1-2 mutants displayed a gain-of-function phenotype (i.e., insensitivity to ABA and cPTIO treatments during seed germination and post-germinative development) was further corroborated in several pANAC089:ANAC089ΔC-1-GFP expression lines, which exhibited ABA-insensitive phenotypes and constitutive nuclear localization (Figures S3A–S3C).
To further characterize the function of the ANAC089 gene in abiotic stress responses, we analyzed seed germination and early development under salt, osmotic, and cold stresses (Figures 1F, 1G, and S3D–S3F). gap1 mutants Col-0 and abi5-1 were sowed on MS medium supplemented with NaCl (100 and 150 mM) and mannitol (250 mM) (Figure 1F). The inhibitory effects of NaCl and mannitol on the germination of gap1 mutants were scored after 7 days of incubation based on green cotyledons and compared to the one in wild-type Col-0 and abi5-1 seeds (Figures 1F and S3D). Similarly, the gap1-1 mutant showed an increased tolerance to freezing temperatures before (Figure S3E) and after cold acclimation (Figures 1G and S3F). These tolerant phenotypes of gap1 gain-of-function mutants to inhibitory conditions for seed germination and seedling establishment paved the way for the ANAC089 regulation of abiotic stress responses.
Differential gene expression pattern in gap1-2 during seed germination
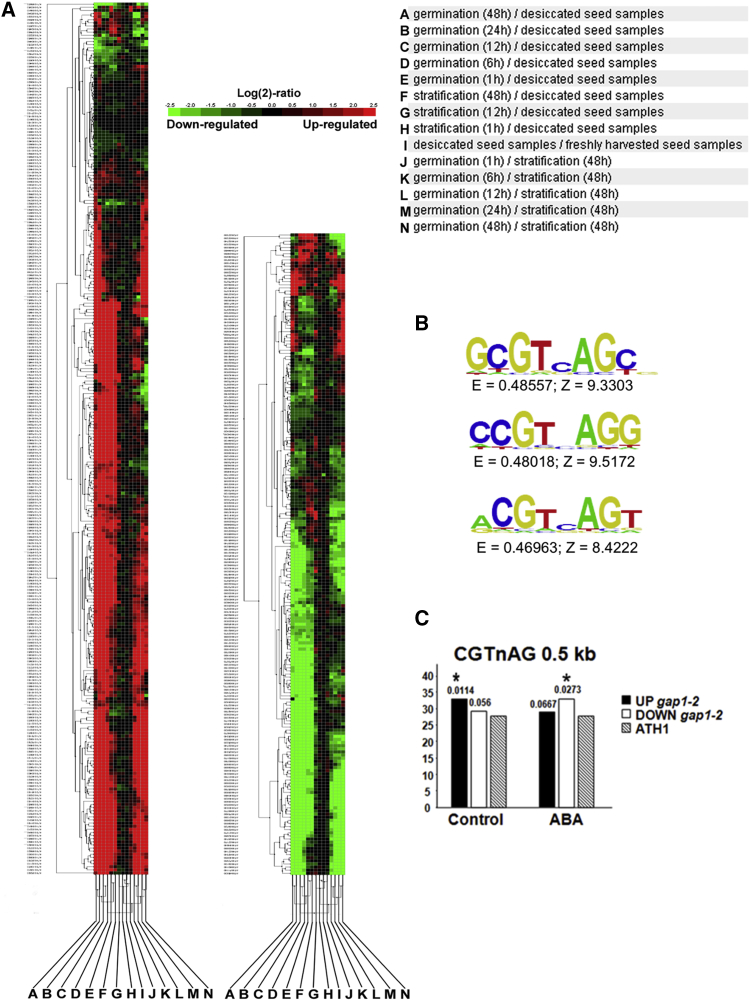
To deepen our understanding of the NO- and ABA-related responses affected in gap1-2 seeds, transcriptional profiling was performed. Transcripts from 3-day-old stratified gap1-2 and Col-0 wild-type seeds after 3 h of imbibition under light were compared, and the results are summarized in Figure 2 (raw data can be found in Tables S1 and S2). Two independent criteria (false discovery rate [FDR] < 0.01 and log2 ratio >1 or <−1) have been followed to select statistically significant differences in gene expression. A hierarchical clustering analysis of 465 genes differentially expressed in gap1-2 versus Col-0, with those regulated by different seed germination, stratification, and desiccation conditions allowed their categorization in different groups (Figure 2A). The most relevant group showed genes related to the start of germination, which were upregulated in gap1-2 mutant versus Col-0 (Figure 2A, left), including genes of cell wall remodeling enzymes and redox homeostasis-related genes. The second group comprises genes downregulated in gap1-2 versus Col-0 that were also repressed in the different comparisons, highlighting ABA and gibberellin (GA) hormone homeostasis (Figure 2A, right).
Figure 2.
Differential transcriptomic profile in gap1-2 during seed germination and consensus cis-element binding site of ANAC089
(A) Hierarchical cluster analysis of 465 differentially expressed ANAC089-responsive genes and at least one of the developmental stages of wild-type (Col-0) Arabidopsis seeds (≥2- and ≤−2-fold change in expression compared to wild type). Genes (rows) and experiments (columns) were clustered by TIGR (The Institute for Genomic Research) using different seed germination experiments. Gene subclusters of interest are discussed in the text.
(B) DNA-binding sequences that are recognized by the ANAC089 protein.
(C) Sequence significantly overrepresented in the promoters of upregulated (control) or downregulated (ABA) genes of ANAC089 in gap1-2 mutant background.
We analyzed the response between upregulated and downregulated genes (265 and 200 genes, respectively, cutoff 2-fold) in gap1-2 mutant versus Col-0 wild-type. ANAC089 gene (fold change = −29.93) is the most strongly repressed gene. This result is consistent with an autoregulatory feedback loop (Figure S1G) and may be explained due to the fact that 6 of the 11 probes of ANAC089 gene, in the Affymetrix GeneChip Arabidopsis ATH1 Genome Array, are integrated inside the deletion present in the ANAC089 of gap1-2 allele (Figures S4A and S4B). Among the 265 genes with increased expression in the gap1-2 mutant, ∼10% are directly related to seed germination processes (Table S1), being the 10 most induced critical genes for cell wall remodeling enzymes (i.e., ATEXPA2, ATEXPA9, XTH19, BXL2, EXP3, and ATPME3). Recently, it has been shown that the TFs NAC25 and NAC1L, both expressed in the seed endosperm, are indispensable for EXPA2 induction by GAs (Sánchez-Montesino et al., 2019). In addition, the GA3ox2 gene responsible for active GA biosynthesis was induced in the gap1-2 mutant, suggesting increased levels of bioactive GAs to promote germination (Finch-Savage and Leubner-Metzger, 2006). Interestingly, ∼8% of the total induced genes belonged to redox-related processes involved in the glutathione and ascorbate cycle and oxidative stress (Table 1) (i.e., l-ascorbate oxidases At5g21105, At5g39580, At2g22420; l-ascorbate peroxidase At1g07890; glutathione peroxidase At2g31570 and glutathione transferases At1g69920, At1g27130, At2g30870, At1g69930). Finally, ∼5% of the upregulated genes were related to cysteine-type endopeptidases and serine-type carboxypeptidases, a group of peptidases previously noted to be involved in ANAC089 cleavage from the membrane (Klein et al., 2012) (i.e., SCPL20, SCPL44, SCPL29, SCPL25, SLP2, CP1, and DELTA-VPE).
Table 1.
Significant upregulated and downregulated genes in gap1-2 mutant versus Col-0 wild type falling in the GO fine category of “redox-related”
| Fold change | Transcript ID | Gene symbol/gene description | CGTnAG 1 kb |
|---|---|---|---|
| 4.78 | At1g26410 | unknown protein similar to reticuline oxidase-like protein; FAD-binding Berberine family protein; oxidoreductase activity | 666 |
| 4.3 | At1g26420 | hypothetical protein similar to reticuline oxidase-like protein; FAD-binding Berberine family protein; oxidoreductase activity | 977 |
| 3.79 | At5g21105 | plant l-ascorbate oxidase; oxidoreductase activity, l-ascorbate oxidase activity, copper ion binding | – |
| 3.33 | At5g20250 | DIN10 (DARK INDUCIBLE 10); hydrolase, hydrolyzing O-glycosyl compounds | 857 |
| 3.1 | At5g39580 | peroxidase ATP24a | 410–983 |
| 3.08 | At5g57220 | CYP81F2; electron carrier/heme binding/iron ion binding/monooxygenase/oxygen binding | – |
| 3.05 | At4g01610 | cathepsin B-like cysteine protease, putative similar to GI: 609175 from Nicotiana rustica | – |
| 2.77 | At5g39610 | ATNAC6 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 6); protein heterodimerization/homodimerization/transcription factor | 214–342 |
| 2.46 | At1g26380 | hypothetical protein similar to reticuline oxidase-like protein GB: CAB45850 GI: 5262224 from Arabidopsis thaliana FAD-binding Berberine family protein; electron carrier activity, oxidoreductase activity, FAD binding, catalytic activity | – |
| 2.42 | At1g69920 | ATGSTU12 (GLUTATHIONE S-TRANSFERASE TAU 12); glutathione transferase | 201 |
| 2.38 | At1g27130 | ATGSTU13 (A. THALIANA GLUTATHIONE S-TRANSFERASE TAU 13); glutathione transferase | – |
| 2.35 | At2g24150 | HHP3/HHP3 (heptahelical protein 3); receptor | 922 |
| 2.3 | At2g30870 | GSTF10 (GLUTATHIONE S-TRANSFERASE PHI 10); copper ion binding/glutathione binding/glutathione transferase | – |
| 2.28 | At5g20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding/electron carrier | – |
| 2.28 | At1g07890 | APX1 (ascorbate peroxidase 1);l-ascorbate peroxidase | 731 |
| 2.18 | At1g60660 | CB5LP (CYTOCHROME B5-LIKE PROTEIN); heme binding | 903–983 |
| 2.17 | At1g69930 | ATGSTU11 (GLUTATHIONE S-TRANSFERASE TAU 11); glutathione transferase | 327 |
| 2.08 | At1g73120 | hypothetical protein; response to oxidative stress | 846 |
| 2.05 | At2g22420 | putative peroxidase; peroxidase superfamily protein; peroxidase activity, heme binding; oxidation reduction, response to oxidative stress | 13 |
| 2.04 | At4g02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | – |
| 2.02 | At2g31570 | ATGPX2 (GLUTATHIONE PEROXIDASE 2); glutathione peroxidase | – |
| −29.93 | At5g22290 | ANAC089 (Arabidopsis NAC domain containing protein 89); transcription factor | – |
| −4.5 | At1g62180 | APR2 (5-ADENYLYLPHOSPHOSULFATE REDUCTASE 2); adenylyl-sulfate reductase/phosphoadenylyl-sulfate reductase (thioredoxin) | 893 |
| −3.25 | At2g29490 | ATGSTU1 (GLUTATHIONE S-TRANSFERASE TAU 1); glutathione transferase | – |
| −2.82 | At4g21990 | APR3 (APS REDUCTASE 3); adenylyl-sulfate reductase | 102 |
| −2.68 | At2g40300 | ATFER4 (ferritin 4); binding/ferric iron binding/oxidoreductase/transition metal ion binding | 992 |
| −2.67 | At1g71695 | peroxidase ATP4a identical to GB:CAA67309 GI:1429213 from A. thaliana | 124 |
| −2.53 | At1g10370 | ERD9 (EARLY-RESPONSIVE TO DEHYDRATION 9); glutathione transferase | – |
| −2.5 | At5g64240 | AtMC3 (metacaspase 3); cysteine-type endopeptidase | – |
| −2.43 | At1g75270 | DHAR2 (DEHYDROASCORBATE REDUCTASE 2); glutathione binding/glutathione dehydrogenase (ascorbate) | 217–656 |
| −2.41 | At4g20820 | reticuline oxidase-like protein reticuline oxidase (EC 1.5.3.9) precursor, Eschscholzia californica; FAD-binding Berberine family protein; oxidation reduction | – |
| −2.36 | At1g20630 | CAT1 (CATALASE 1); catalase | 540 |
| −2.22 | At2g29420 | ATGSTU7 (A. THALIANA GLUTATHIONE S-TRANSFERASE TAU 7); glutathione transferase | – |
| −2.05 | At1g75280 | NADPH oxidoreductase, putative similar from A. thaliana | 736 |
Genes with specific cis-acting elements recognized by the ANAC089 TF are highlighted in bold. GO, Gene Ontology; GI, Gene Identifier; GB, Gene Bank.
Concerning the 200 genes detected with repressed expression in the microarray (Table S2), apart from ANAC089 and NITRILASE4 (NIT4) (Figure S4C) as the highest repressed genes, one of the most important genes involved in the induction of seed dormancy, DOG1, was strongly repressed (fold change = −5.12). DOG1 expression and especially DOG1 protein levels directly determine the dormancy release of freshly harvested seeds (Nakabayashi et al., 2012). This pronounced DOG1 repression may cooperate with the NO effect in the non-dormant phenotype displayed by the gap1-2 mutant (Figure 1D). In addition, significant changes in ABA and GAs homeostasis-related genes were found (Figure S4D), supporting the ABA insensitivity displayed in the gap1-2 mutant, mainly NCED9 from the ABA biosynthetic pathway (Frey et al., 2012; Kanno et al., 2010), GA2ox2, one of the main genes responsible for GA catabolism (Thomas et al., 1999), and GA20ox3 of the GA biosynthetic pathway (Phillips et al., 1995). Moreover IAA30, previously reported to be an inhibitor of seed germination under salt stress conditions after induction by the NTM2 (ANAC069) (Park et al., 2011), was also repressed in the gap1-2 mutant background. As shown in Table 1 and Figure S4E, redox-related genes (6% of the total repressed genes) had an altered expression in gap1-2 (i.e., glutathione transferases At2g29490, At1g10370, and At2g29420, and glutathione dehydrogenase DHAR2 and catalase CAT1). Therefore, the altered expression of hormone and redox-related genes (either induced or repressed) in gap1-2 mutant seeds may be responsible for the changes in cell redox status during germination.
DNA binding specificity of the ANAC089 TF
To identify the consensus cis-element binding site of ANAC089, we have used a DNA protein binding microarray (PBM) (Godoy et al., 2011). The sequence significantly overrepresented in the PBM analysis was GCGTCAGC and the two variants CCGTCAGG and ACGTCAGT (Figure 2B).
The complete DNA-binding sequence, which is recognized by the ANAC089 protein, includes modifications to the so-called NAC RECOGNITION SEQUENCE (NACR) CATGTG and the NAC-BINDING SITES (NACBS) CGTGA (Jensen et al., 2010; Olsen et al., 2004, 2005; Tran et al., 2004). As shown in Figure 2C, Tables 1, S1, and S2, sequence examination revealed the CGTnAG motifs in the promoter region of 47% (28% upregulated and 19% downregulated) of the total differentially expressed genes. Furthermore, this CGTnAG motif was overrepresented in upregulated genes of the gap1-2 mutant background in the microarray performed in control conditions (Figure 2C). Conversely, this consensus cis-element binding site of ANAC089 was overrepresented in the downregulated genes of the gap1-2 mutant background in a microarray performed after 3 h of 5 μM ABA treatment (Figure 2C and GEO: GSE25159). All of these results indicate the ability of ANAC089 to discriminate and bind a specific cis-regulatory element in the promoters of redox homeostasis and ABA-responsive genes, respectively, to regulate Arabidopsis seed germination.
gap1 mutants alter NO, ABA, thiol, and disulfide levels in seeds
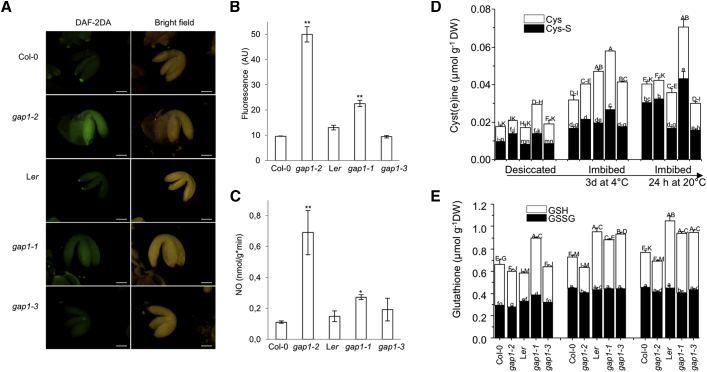
We further analyzed the extent to which the transcriptional changes may reflect hormone and metabolic alterations during seed germination. To this end, we examined the endogenous NO levels in 24-h-imbibed seeds and isolated embryos by using 2 independent methods (Mur et al., 2011), the fluorescence indicator 4,5-diaminofluorescein diacetate (DAF-2DA) and the Griess assay (Liu et al., 2009; Fernández-Marcos et al., 2011; Albertos et al., 2015). The NO-specific dye reveals higher levels of NO abundance in gap1-1 and gap1-2 mutants after seed imbibition and localized mainly in the seed endosperm, as described previously (Liu et al., 2009), but also in seed embryos (Figures 3A, 3B, S5A, and S5B). Application of the NO scavenger cPTIO reduced the NO-dependent DAF-2DA fluorescence signal (Figures S5C and S5D). In addition, the NO release during seed imbibition measured by the Griess assay in gap1-2 and gap1-1 backgrounds was increased almost 6-fold and 2-fold compared with wild-type Col-0 and Ler, respectively (Figure 3C). Examination of endogenous NO levels were also analyzed in ABA-treated seeds with DAF-2DA (Figures S5E and S5F). In control conditions, gap1-1 and gap1-2 showed constitutively high NO levels that slightly increased after ABA treatment. However, in wild-type seeds from Col-0 and Ler, the NO levels after ABA treatment were highly increased. These results confirmed that the gain-of-function mutants had already constitutive higher NO levels and were partially insensitive to the effect of ABA. Meanwhile, the wild-type clearly responded to an ABA treatment that prompted endogenous NO production, as previously described (Desikan et al., 2002; Planchet et al., 2014).
Figure 3.
Endogenous NO levels in embryos and total thiols and disulfides in seeds
(A) Fluorescence corresponding to NO accumulation in embryos 24 h after seed imbibition. Scale bars, 100 μm.
(B) Quantitative data of NO-dependent DAF-2DA fluorescence embryo images. Values represent the means ± SEs (n = 3). Asterisks indicate statistically significant differences between gap1-2 versus Col-0 (t test, ∗∗p < 0.01); gap1-1 versus Ler (t test, ∗p < 0.05).
(C) Measurement of NO release (nmol/g ∗ min) after 24 h in wild-type (Col-0, Ler) and mutant (gap1-1, gap1-2, gap1-3) seeds. Error bars represent means ± SEs (n = 3). Asterisks indicate statistically significant differences between gap1-2 versus Col-0 (t test, ∗∗p < 0.01); gap1-1 versus Ler (t test, ∗p < 0.05).
(D and E) Levels of cystine (black bars) and cysteine (white bars) (D) and glutathione disulfide (GSSG, black bars) and glutathione (GSH, white bars) (E) in after-ripened, dry, and imbibed seeds of gap1-1 and gap1-2 mutants compared to the wild-type (Col-0, Ler) and gap1-3 mutant. Values represent means ± SEs (n = 3). Letters indicate statistically significant differences (p < 0.05) according to 1-way ANOVA with Tukey’s post hoc test.
Furthermore, ABA endogenous levels in desiccated and 24-h-imbibed seeds were also quantified (Figure S5G). A clear decrease in ABA content was found in gap1-1 and gap1-2, whereas it was not apparent in gap1-3. The major significant difference in ABA content between the desiccated and imbibed seed batches is reliable for gap1-1 versus Ler. Accordingly, gap1 gain-of-function mutants display the insensitive ABA and NO-depletion (caused by cPTIO during seed germination) phenotypes, as a consequence of their lower ABA and higher NO endogenous levels after seed imbibition. Increasing NO levels by exogenous application of NO donors provoke seed dormancy breaking and germination promotion (Beligni and Lamattina, 2000; Bethke et al., 2004a, 2004b).
We previously investigated the ABA and NO crosstalk and the effect of altered NO levels in the ABI5 protein accumulation during seed germination (Albertos et al., 2015). The ABI5 central repressor is S-nitrosated by the NO produced after seed imbibition, promoting the interaction with CUL4-based and KEG E3 ligases, and it is rapidly degraded by the proteasome during seed germination. Since the 2 gap1 isolated mutants display elevated endogenous NO levels after seed imbibition, we analyzed whether the ABI5 protein levels were modulated in gap1 mutant backgrounds after seed imbibition. We observed a slight decrease in ABI5 levels in the gap1-2 mutant and a notable decrease in the gap1-1 mutant background regarding their respective wild-type, Col-0, and Ler (Figures S5H and S5I).
In addition, low-molecular-weight (LMW) thiols and disulfides were measured in after-ripened seeds. In dry seeds, there were significant changes in the level of total (thiol and disulfide) cysteine and total glutathione, in which gap1-2 had more total cyst(e)ine than Col-0 and gap1-1 had more total cyst(e)ine and total glutathione than Ler; gap1-3 had wild-type levels of glutathione and cysteine (Figures 3D and 3E). This consistent difference was also revealed in the half-cell redox potential analysis, but only in the Ler background. Therefore, these mutations affect seed development on the maternal plant in a consistent way, which could also affect germination-related processes. After stratification (3 days at 4°C), a significant accumulation of cysteine occurred in all of the genotypes, which continued during germination (24 h at 20°C), whereby gap1-1 had double the cysteine levels relative to Ler, indicating that stratified seeds are at different physiological states/germination stages. The significantly higher levels of the disulfide cysteine in gap1-1 could be explained by increased NO production in these mutants, which could oxidize LMW thiols to disulfides. Changes in the thiol:disulfide ratio led to alterations in the GSNO concentration, the main endogenous NO storage (Malik et al., 2011; Mur et al., 2013), increasing NO levels during seed germination.
Subcellular localization of ANAC089 is altered by redox modifications
To determine the possible effects of NO-, reactive oxygen species (ROS)-, and redox-related compounds in the subcellular localization of ANAC089 protein, we analyzed the ROS content of wild-type plants and gap1 mutants with the fluorescence probe 2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA) (Figures S6A–S6C). The dominant mutants gap1-1 and gap1-2 showed more reduced ROS content than wild-type plants and the knockout mutant gap1-3. This feedback regulation between the levels of NO and ROS in the plant has been already reported by Sanz et al. (2014) and reviewed in Lindermayr and Durner (2015).
Then, we generated Arabidopsis transgenic lines by expressing GFP-ANAC089 under control of the 35S promoter in the wild-type (Col-0) background (Figures 4, S6D, and S6E). This line showed a wild-type phenotype under ABA treatment (Figure S6D), suggesting that the GFP tag in the N-terminal part of ANAC089 affected the functional properties of the TF (i.e., DNA binding), as previously reported by Li et al. (2011).
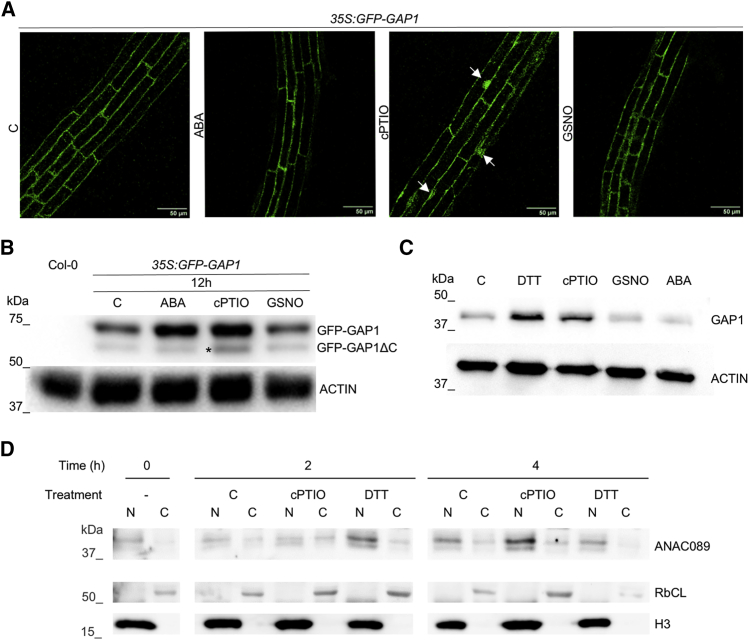
Figure 4.
ANAC089 subcellular localization and protein accumulation in response to NO- and redox-related treatments
(A) Confocal microscopy of 7-day-old Arabidopsis roots of 35S:GFP-ANAC089-overexpressor lines after 4 h of control (C), 5 μM ABA, 1 mM cPTIO, and 1 mM GSNO treatments. Nuclear localization of GFP-ANAC089 protein after the corresponding treatments is indicated by arrows. Scale bars, 50 μm.
(B) Immunoblot analysis of GFP-ANAC089 protein levels in 7-day-old seedling extracts of 35S:GFP-ANAC089-overexpressor line under control (C) conditions or treated 12 h with 5 μM ABA, 500 μM cPTIO, and 500 μM GSNO. Asterisk (60 kDa) indicates nuclear GFP-ANAC089ΔC localized protein while the extranuclear GFP-ANAC089 protein is 65 kDa. Actin protein levels are shown as a loading control.
(C) ANAC089 levels in 3-day-old stratified Col-0 seed extracts (C) and after 3 h of treatments with 1 mM DTT, 500 μM cPTIO, 500 μM GSNO, and 5 μM ABA. Actin protein levels are shown as a loading control.
(D) Subcellular localization of ANAC089 protein by nuclear/cytoplasmic fractioning assays in 7-day-old 35S:GFP-ANAC089 Arabidopsis seedlings grown in MS medium (C) or MS medium supplemented with 1 mM cPTIO and 1 mM DTT for 0, 2, and 4 h. Histone3 (H3) and Rubisco large subunit (RbcL) were used as loading controls from nucleus (N) and cytoplasm (C), respectively.
Previous confocal microscopy studies located the ANAC089 full version in the cytoplasm for in planta studies on Arabidopsis or in the endoplasmic reticulum for in vivo studies on Arabidopsis protoplasts and Nicotiana benthamiana leaves (Klein et al., 2012; Li et al., 2010, 2011; Smyczynski et al., 2006). Truncated versions of ANAC089 lacking the TM present constitutive nuclear localization (Klein et al., 2012; Li et al., 2010, 2011; Yang et al., 2014). Thus, 35S:GFP-ANAC089-overexpressor lines were studied using confocal laser scanning microscopy to examine the localization of the GFP-ANAC089 in planta under different NO/redox-related and ABA treatments. GFP-ANAC089 displayed extranuclear localization in control conditions corroborated by previous investigations (Li et al., 2011; Yang et al., 2014) (Figure 4A). This subcellular localization was also detected after cold treatment (4°C), ABA, NO-donor GSNO, and glutathione disulfide (GSSG) (Figures 4A and S6F). Conversely, treatments with the NO scavenger cPTIO and with reductant agents, such as dithiothreitol (DTT) and glutathione (GSH), released GFP-ANAC089 from the C-terminal TM anchored to the membrane to allow the nuclear subcellular localization of the protein (Figures 4A and S6F).
To extend these findings, the chimeric GFP-ANAC089 and the wild-type ANAC089 proteins under different treatments were analyzed by western blot (Figures 4B and 4C). Seven-day-old GFP-ANAC089-overexpressor lines were untreated (C) or treated for 12 h with 5 μM ABA, 500 μM cPTIO, and 500 μM GSNO before collection for western blot analysis. Seedlings treated with cPTIO resulted in higher GFP-ANAC089 levels and increased truncated/nuclear versions of GFP-ANAC089ΔC lacking the TM, with 5 kDa less than the GFP-ANAC089 (Figure 4B lower band, asterisk). Likewise, 3-day-old stratified Col-0 seeds were untreated (C) or treated for 3 h with 5 μM ABA, 500 μM cPTIO, 500 μM GSNO, and 1 mM DTT, and wild-type ANAC089 protein levels were analyzed. Again, an increase in ANAC089 levels was found exclusively after treatments with cPTIO and the reductive agent DTT (Figure 4C).
To further confirm that the signal triggered the cleavage and translocation of ANAC089 protein to the nucleus occur in a short window, a subcellular fractionation was carried out (Figure 4D). Thus, 7-day-old 35S:GFP-ANAC089 seedlings were treated with DTT or cPTIO or non-treated for 2 and 4 h, and the nucleus and the cytoplasm were fractionated. After 2 h of treatment with DTT, there was an enrichment of ANAC089 in the nuclear fraction, while under the cPTIO scavenger, this enrichment appeared after 4 h of treatment. These findings imply that alterations in the cell redox status, either by decreasing endogenous NO levels (cPTIO) or increasing reductant agents (DTT), translocate ANAC089 into the nucleus and promote ANAC089 protein accumulation.
Discussion
The plant-specific family of NAC TFs is involved in myriad developmental programs, hormonal signaling, and biotic and abiotic stress responses (Nakashima et al., 2012; Puranik et al., 2012). The conserved NAC domain within the N-terminal region harbors the DNA binding domain, the dimerization domain, and the nuclear localization sequence (Ernst et al., 2004). However, high variability in the C-terminal part of the NAC TFs is exhibited, including the presence of a TM (Ooka et al., 2003). ANAC089 contains all of these features and has been sorted in the group of OsNAC08-related NACs together with ANAC060 and ANAC040 (Ooka et al., 2003).
The membrane-tethered ANAC089 protein shows extranuclear localization, requiring detachment after particular and specific signals to reach the nucleus and regulate gene expression (Klein et al., 2012; Li et al., 2010, 2011; Yang et al., 2014). Hence, the lack of the membrane association domain causes the inability of ANAC089 to anchor into the membrane, and consequently, the truncated proteins display constitutive nuclear localization exhibiting major functions in different processes—for example, suppressing fructose signaling (Li et al., 2011) and promoting endoplasmic reticulum stress-induced programmed cell death (Yang et al., 2014). The premature stop codon mutation in gap1-1 and the deletion in the C-terminal region in gap1-2 result in truncated proteins lacking the membrane-bound domain, as detected in the imbibed seeds of these mutants. Thus, the presence of C terminally truncated versions of ANAC089 protein is responsible for the gain-of-function phenotypes of gap1-1 and gap1-2 mutants in ABA and cPTIO during seed germination. These truncated ANAC089 proteins (ANAC089ΔC-1 and ANAC089ΔC-2) are constitutively located in the nucleus and regulate essential ABA responsive and redox-related homeostasis genes, placing ANAC089 as a central regulator of myriad abiotic stress (i.e., salt, osmotic, and cold) responses during Arabidopsis seed germination and post-germinative development. Accordingly, the transgenic lines expressing complete versions of ANAC089 protein with the GFP tag fused to the N-terminal show endoplasmic reticulum localization that maintains the protein outside the nucleus, causing wild-type responses to the stimulus analyzed (Li et al., 2011 and the present work). It has been reported that ANAC060 expression is induced by ABI4, belonging to the sugar-ABA signaling cascade, but the nuclear ANAC060 lacking the TM attenuates ABA induction and ABA signaling, resulting in sugar insensitivity during seed germination (Li et al., 2014). Similarly, our evidence supports that ABA can induce the expression of ANAC089 and protein accumulation in seeds and seedlings.
The freezing tolerance phenotype of 2-week-old gap1-1 mutants before and after cold acclimation confers a notable role onto ANAC089 during cold stress response. Several tethered membrane ANAC proteins are regulated under cold or heat stress conditions (Kim et al., 2007; Seo et al., 2010). The integration of the ABA, fructose, and redox-related signaling pathways with this noteworthy function of ANAC089 remains to be investigated. Of note, NO production has been involved in cold acclimation and freezing tolerance in Arabidopsis (Costa-Broseta et al., 2018, 2019; Zhao et al., 2009).
Transcriptomic profiling revealed differentially expressed genes in the gap1-2 mutant background crucially related to germination processes such as ABA and GA hormone metabolism, modifying the cellular redox status. To detect cis-regulatory sequences, we used high-throughput identification of the consensus binding site of ANAC089 to search for overrepresented motifs in the promoters of ANAC089 differentially expressed genes, considering that co-regulated genes usually depend on the same TF for their expression. Thus, ANAC089 has the ability to bind the specific cis-regulatory CGTnAG motif that is overrepresented in the promoters of upregulated redox-related homeostasis genes and downregulated ABA-responsive genes.
Several authors reported the tight relationship between ANAC089 and redox status (Klein et al., 2012; Yang et al., 2014). On the one hand, treatments with the reducing agent DTT and tunicamycin, which promote protein misfolding and induces endoplasmic reticulum stress, respectively, release ANAC089 from the trans-Golgi network and endoplasmic reticulum membrane and relocate it in the nucleus (Klein et al., 2012; Yang et al., 2014). On the other hand, this nuclear relocated ANAC089 protein negatively regulates the expression of sAPX (stromal ASCORBATE PEROXIDASE) and plays an important role in regulating downstream genes involved in programmed cell death (Klein et al., 2012; Yang et al., 2014). We provide significant evidence that ANAC089 has essential roles in the regulation of redox-related homeostasis genes identified through the microarray analysis. Remarkably, some of these genes also contain the specific cis-regulatory sequence for ANAC089 binding in their promoter region. Thus, first, the modification of cell redox status directed by ANAC089 may be responsible for displaying increased NO levels in gap1-1 and gap1-2 mutants in imbibed seeds and cPTIO-insensitive phenotype during germination. Second, this redox status, with a reduction in ROS levels, may negatively affect the membrane-anchoring domain, increasing the rate of non-anchoring protein that may be processed and translocated into the nucleus to activate the response against ABA and/or stress. Furthermore, different treatments with NO- and redox-active compounds such as cPTIO and GSH reveal changes in ANAC089 subcellular localization in planta, and release ANAC089 to the nucleus, highlighting again the feedback interaction between the cell redox status and ANAC089 reported previously (Klein et al., 2012; Yang et al., 2014). Apart from the effect on the subcellular localization, changes in ANAC089 accumulation under treatments with DTT and cPTIO were detected, suggesting that ROS and/or NO balance may regulate the stability of ANAC089. We therefore conclude that redox-sensitive ANAC089 integrates ABA and NO responses during seed germination and early development in Arabidopsis, conferring key functions to this hub/central regulator in the successful growth of the plant under abiotic stress.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Living Colors® GFP Monoclonal Antibody | Clontech | Cat#632375; RRID:AB_2756343 |
| Anti-ANAC089 Purified Rabbit Immunoglobulin | Biomedal | Custom antibody |
| Anti-ABI5 Purified Rabbit Immunoglobulin | Biomedal | Custom antibody |
| Anti-Actin clone 10-B3 Purified Mouse Immunoglobulin | Sigma-Aldrich | Cat#A0480; RRID:AB_476670 |
| ECL Rabbit HRP-linked | GE Healthcare Life Sciences | Cat#NA934;RRID: AB_772206 |
| ECL Mouse IgG, HRP-linked | GE Healthcare Life Sciences | Cat#NA931; RRID:AB_772210 |
| Anti-Maltose Binding Protein antibody | Abcam | Cat#ab9084; RRID:AB_306992 |
| (Goat) anti-rabbit IgG DyLight 549 conjugated | Pierce | Cat#DI-1549; RRID:AB_2336407 |
| H3 | Histone H3 (rabbit antibody) (nuclear marker) | Agrisera | Cat#AS10 710; RRID:AB_10750790 |
| RbcL | Rubisco large subunit | Agrisera | Cat#AS03 037; RRID:AB_2175402 |
| Bacterial and virus strains | ||
| BL21(DE3) Competent E. coli | NEB | Cat#C2527I |
| One-Shot TOP10 E. coli | Invitrogen | Cat#K240020 |
| Agrobacterium tumefaciens strain C58C1 (pGV2260) | Deblaere et al., 1985 | N/A |
| Agrobacterium tumefaciens strain GV3101 | Nambara Lab | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Abscisic acid, ABA | Sigma-Aldrich | Cat#A1049 |
| Carboxy-PTIO | Thermofisher | Cat#C7912 |
| Sodium chloride | Sigma-Aldrich | Cat#S7653 |
| Mannitol | Sigma-Aldrich | Cat#M4125 |
| 4,5-Diaminofluorescein diacetate solution, DAF-2DA | Sigma-Aldrich | Cat#D225 |
| S-Nitrosoglutathione, GSNO | Sigma-Aldrich | Cat#N4148 |
| L-Glutathione oxidized, GSSG | Sigma-Aldrich | Cat#G4376 |
| L-Glutathione reduced, GSH | Sigma-Aldrich | Cat#G4251 |
| DL-Dithiothreitol | Sigma-Aldrich | Cat#D0632 |
| Bleach (4-5% sodium hypochlorite) | Conejo | N/A |
| Triton 100X | Sigma-Aldrich | Cat#T8787 |
| MURASHIGE & SKOOG MEDIUM + VITAMINS/MES | Duchefa Biochemie | Cat#M0255.0001 |
| Sucrose | Sigma-Aldrich | Cat#S7903 |
| PLANT AGAR | Duchefa Biochemie | Cat#P1001 |
| Potassium hydroxide | Sigma-Aldrich | Cat#P1767 |
| Recombinant protein ANAC089 | Biomedal | Custom r. protein |
| Trizma® base | Sigma-Aldrich | Cat#T1503 |
| Sodium chloride | Sigma-Aldrich | Cat#S7653 |
| EGTA | Sigma-Aldrich | Cat#E3889 |
| Magnesium chloride | Sigma-Aldrich | Cat#M8266 |
| Sodium fluoride | Sigma-Aldrich | Cat#S7920 |
| β-Glycerol phosphate | Sigma-Aldrich | Cat#50020 |
| Immobilon-P Membran, PVDF, 0,45 μm | Millipore | Cat#IPVH00010 |
| Sodium pyrophosphate tetrabasic | Sigma-Aldrich | Cat#P8010 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#04693124001 |
| Bio-Rad Protein Assay | Bio-Rad | Cat#5000001 |
| Tween® 20 | Sigma-Aldrich | Cat#P2287 |
| Critical commercial assays | ||
| Nitric Oxide (total), detection kit (Griess assay) | Enzo Life Sciences | Cat#ADI-917-020 |
| RNeasy Plant Mini Kit | QIAGEN | Cat#74904 |
| CelLytic PN Isolation/Extraction Kit | Sigma-Aldrich | Cat#CELLYTPN1 |
| ECL Advance Western Blotting Detection Kit | GE Healthcare Life Sciences | Cat#RPN2135 |
| Experimental models: organisms | ||
| Arabidopsis thaliana lines | ||
| Col-0 | NASC | N1093 |
| Ler | NASC | NW20 |
| gap1-1 | This study | N/A |
| gap1-2 | This study | N/A |
| gap1-3 | CSHL | CSHL_GT19225 |
| abi5-1 | NASC | N8105 |
| pGAP1:GAP1ΔC1-GFP | This study | N/A |
| 35S:GFP-GAP1 | This study | N/A |
| Nicotiana benthamiana | N/A | N/A |
| M2 seeds: EMS-Col, EMS-Ler and FN-Col | Lehle Seeds | N/A |
| Oligonucleotides | ||
| Primers | N/A | See Table S3 |
| Recombinant DNA | ||
| pENTR/D-TOPO | Invitrogen | Cat#K240020 |
| pENTR/D-TOPO-pANAC089:ANAC089ΔC-1 | This study | N/A |
| pGWB4 | T. Nakagawa, Shimane U. | N/A |
| pGWB4-pANAC089:ANAC089ΔC-1 | This study | N/A |
| pENTR/D-TOPO-CaMV35S:ANAC089-GFP | This study | N/A |
| pMDC45 | M. Curtis, U. Grossniklaus | Cat#CD3-739 |
| pMDC45-CaMV35S:ANAC089-GFP | This study | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Office Excel Power Point 2016 | Microsoft | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oscar Lorenzo (oslo@usal.es).
Materials availability
Plant lines generated in this study will be made available on request, but we may require a completed Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
The published article includes all raw and analyzed datasets generated during this study. Original data have been deposited to NCBI GEO: GSE25159. Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession number: NAC089, AT5G22290.
Experimental model and subject details
Plant material
Arabidopsis thaliana (L.) Heynh ecotypes Columbia (Col-0) and Landsberg erecta (Ler) are the genetic backgrounds for all wild-type plants used in this work. gap1-1 (bck Ler) and gap1-2 (bck Col-0) mutant plants were genuinely generated and identified for this work. Seed stock of anac089-3 loss of function allele was obtained from the Martienssen laboratory at CSHL Arabidopsis gene trap collection lines (http://genetrap.cshl.edu). Genetic crosses were performed using both gap1-2 and Col-0 as donor and acceptor, respectively. F1 seeds were sown in 1μM ABA to check dominant mutation (Figure S1D). Arabidopsis thaliana pGAP1:GAP1ΔC1-GFP and 35S:GFP-GAP1 transgenic plants were generated for this work by floral-dipping Col-0 plants (Clough and Bent, 1998) with Agrobacterium tumefaciens containing pGWB4-pGAP1:GAP1ΔC1-GFP and pMDC45-35S:GFP-GAP1 plasmid, respectively.
Growth conditions
For in vitro culture, Arabidopsis seeds were surface-sterilized in 75% (v/v) bleach (4%–5% sodium hypochlorite) and 0.01% (w/v) Triton X-100 for 5 min and washed three times in sterile water before sowing. Seeds were stratified for 3 d at 4°C and then sowed on Murashige and Skoog (MS) (Murashige and Skoog, 1962) solid medium with 2% (w/v) Suc and 0.6% (w/v) agar and the pH was adjusted to 5.7 with KOH before autoclaving. Seeds were sown on the media supplemented or not with different treatments ABA, cPTIO or GNSO and plates were sealed and incubated in a controlled environment growth chamber.
Six- to fifteen-day-old Arabidopsis pGAP1:GAP1ΔC1-GFP and 35S:GFP-GAP1 seedlings were grown in vitro and treated with NO scavenger (cPTIO), NO donor (GSNO) reducing agents (DTT, GSH) or ABA between 2 or 4 hours before imaging with confocal microscopy or nucleus/cytoplasm fragmentation assays.
For seed propagation, Arabidopsis plants were grown in a growth chamber or greenhouse under 50%–60% humidity, a temperature of 22°C and with a 16-h light/8-h dark photoperiod at 80 to 100 μE m-2 s-1 in pots containing 1:3 vermiculite:soil mixture.
Method details
Mutant screening
M2 seeds of ethyl methane sulfonate (EMS)-mutagenized Columbia (EMS-Col), EMS-mutagenized Landsberg erecta (EMS-Ler) and fast neutron-mutagenized Columbia (FN-Col) were purchased from Lehle Seeds (Round Rock, TX) and each of approximately 20,000 M2 seeds (20 batches of ∼1,000 M2 seeds harvested from ∼1,000 M1 seeds) was used for screening to isolate ABA and cPTIO-insensitive mutants. Screening conditions were described previously (Nambara et al., 2002) except for (+)-S-ABA and cPTIO were used. Briefly, M2 seeds were surface-sterilized, sown on 0.8% agar plates supplemented with half strength of MS and 3μM (+)-S-ABA or 100μM cPTIO. Seeds were stratified for 4 days and incubated in the presence of ABA for 4 days at room temperature under continuous light condition. Germination in cPTIO was tested under continuous light conditions at room temperature without stratification. Seedlings were transferred to hormone-free media, incubated for several days and transferred to pots for seed harvest.
Mapping and cloning of the ANAC089 locus
Mutants were crossed with wild-type of different accession from the mutant. SNPs of Col/Ler were used for mapping the mutations. The gap1-1 and gap1-2 mutations were independently mapped using 376 and 491 F2 plants, respectively. Following CAPS markers defined the 86 kb and 25 kb regions of gap1-1 and gap1-2, respectively (Table S3). Genomic DNA corresponding to candidate genes was amplified by PCR from gap1-1 mutant plants and sequenced to identify the gap1 mutation.
Germination assays
To measure ABA and NO sensitivity during germination, seeds were plated on solid medium composed of MS basal salts, 2% (w/v) Suc or 0.6% w/v agar plates (adjusted to pH 5.8 by MES buffer), and different concentrations of 0.5, 1, 3 or 5μM ABA, 100 μM cPTIO (NO scavenger) and/or 500 μM GSNO (NO donor). Seed lots to be compared were harvested at same seed maturation stages from individual plants grown under identical environmental conditions. For the dormancy assays freshly harvested seed lots were directly used without stratification. Seed germination was assessed during 10 days after sowing as the percentage of seeds with an emerged radicle or seedlings that germinated and developed green fully expanded cotyledons. Each value represents the average germination percentage of 50 to 100 seeds with the SE of three replicates. Experiments were repeated at least three times with three independent seed batches obtaining similar results.
Cold stress tolerance assays
For freezing tolerance assays, 2-week-old Arabidopsis plants growing in soil under control conditions (non-acclimated) or exposed 7 additional days to 4°C (cold acclimated) were transferred to a freezing chamber set to 4°C for 30 min in darkness. Subsequently, temperature was allowed to decrease at a rate of – 1°C per 30 min until reaching the final desired freezing temperature, which was maintained for 6 h. Then, temperature was increased to 4°C at the same rate and thawing was allowed for 12 h before returning plants to control conditions for recovering. Tolerance to freezing was determined as the percentage of plants surviving after one week of recovery.
Constructs and transgenic plants
A 4,077-bp fragment of gap1-1 ANAC089 genomic DNA was amplified by PCR using primers “Compl” (Table S3), to obtain the sequence pANAC089:ANAC089ΔC-1. This PCR product was cloned into pENTR/D-TOPO vector (Invitrogen), and transferred into the binary vector pGWB4 (GFP tag at the C terminus) via the LR reaction (Invitrogen) (Nakagawa et al., 2007). To generate CaMV35S:ANAC089-GFP, a 1,022-bp cDNA of ANAC089 was PCR amplified with the primers “cDNA” (Table S3). The PCR product was cloned into pENTR/D-TOPO vector (Invitrogen) and recombined into pMDC45 (GFP tag at the N terminus) using Gateway LR Clonase (Invitrogen). The binary vectors pGWB4 and pMDC45 were introduced into Agrobacterium tumefaciens strain GV3101 and C58C1 (pGV2260), respectively, and used to transform Col-0 plants by floral dip method (Clough and Bent, 1998) as described previously (Saavedra et al., 2010).
Detection of endogenous NO
24 hours stratified seeds of Col-0, Ler, gap1-1, gap1-2 and gap1-3 untreated or treated with 5 μM ABA or 1 mM cPTIO were assayed with 4,5-diaminofluorescein diacetate (DAF-2DA, Sigma) for detection of endogenous NO as previously described (Albertos et al., 2015). Quantification of fluorescence intensity was performed with ImageJ software. To support these results, NO production was measured spectrophotometrically at 540 nm using the Griess reaction within the Total Nitric Oxide Kit (Stressgen) following the manufacturer’s instructions. The final NO concentration expressed in nanomoles per gram per minutes was measured in 50 mg of 24h imbibed seeds of Col-0, Ler, gap1-1, gap1-2 and gap1-3 after the correlation between the total nitrite concentration of the samples and a nitrite standard curve as previously described (Liu et al., 2009; Fernández-Marcos et al., 2011).
Detection of endogenous ROS
ROS levels were detected in 6-day-old Arabidopsis seedlings of Col-0, Ler, gap1-1, gap1-2 and gap1-3 using 2′,7′-dichlorofluorescein diacetate (DCF-DA) at 5 μM in Tris-HCl 10 mM pH 7,4 for 30 min in dark conditions. Fluorescence was examined with a LEICA DM LB Microscope. Excitation source was a mercury lamp EBQ 100 Isolated and fluorescence was detected by a GFP filter. Quantification of intensity was analyzed with ImageJ software. Sample without the DCF-DA was used as a negative control.
Analysis of LMW thiols and disulfides
Low-molecular-weight (LMW) thiols and disulfides were measured using the HPLC method specified in Bailly and Kranner (2011). Briefly, ca. 15 mg freeze-dried seeds (> 50 seeds) / replicate (n = 2-3) were finely ground before being extracted in 1 mL of 0.1 M ice-cold HCl, centrifuged for 20000 g for 10 min at 4°C and 800 μl of supernatant were transferred avoiding the congealed lipids. The extract was re-centrifuged as before and 120 μl of the supernatant was taken for the determination of total thiols and disulfides. First, the pH was adjusted to 8.0 using 180 μl of 200mM bicine buffer (pH 9.1) and disulfides were reduced by DTT before labeling thiol groups with monobromobimane (mBBr) and terminating the reaction with methanesulfonic acid. To measure disulfides only, 400 μl of supernatant was pH-adjusted to 8.0 with 600μl bicine buffer. LMW thiols were blocked with N-ethylmaleimide (NEM) and excess NEM was removed with an equal volume of toluene four times before disulfides were reduced with DTT and labeled with mBBr as above. After further centrifugation, the labeled LMW thiols were separated by reversed-phase using an Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA, USA) on a ChromBudget 120-5-C18 column (5.0 μm, BISCHOFF Analysentechnik u. -geraete GmbH, Leonberg, Germany) and detected by fluorescence (excitation wavelength = 380 nm, emission wavelength = 480 nm). Data were calculated using individual calibration curves for each LMW thiol that were linear over the range measured. The amount of LMW thiols were calculated by subtracting the amount of LMW disulfides from the amount of total LMW disulfides and thiols.
LC-MS/MS analysis of ABA
Analyses of ABA were carried out according to Schausberger et al. (2019). In brief, 10-20 mg of lyophilized seed powder were extracted in 1.5 ml of ice-cold acetone/water/acetic acid (80:20:1, v:v:v) after addition of 25 μl stable isotopically labeled internal standard (IS) solution (1 μM ABA-d6) by shaking (TissueLyser II, QIAGEN, Düsseldorf, Germany) at 30 Hz for 5 min using two 3 mm agate beads per Eppendorf tube, followed by centrifugation at 10,000 × g, 4 °C for 12 min. Supernatants were evaporated to dryness using a SpeedVac SPD111 vacuum concentrator (Thermo Fisher Scientific Inc., Waltham, MA, USA), followed by resuspension in 150 μl of ACN/water (50:50, v:v), supported by 5 min ultrasonication in an ice-cooled water bath. The extracts were filtered through 0.2 μm PTFE filters before injection into the UHPLC-MS/MS system. ABA was identified and quantified by LC-MS, using an ekspert ultraLC 100 UHPLC system coupled to a QTRAP 4500 mass spectrometer (AB SCIEX, Framingham, MA, USA).
Quantitative RT-PCR
Total RNA preparation from Col-0 and gap1-2 mutant seeds and first strand cDNA synthesis were carried out as described previously (Okamoto et al., 2006) and quantitative RT-PCR (Q RT-PCR) was performed by the QuantiTect SYBR Green PCR kit (QIAGEN) as described previously (Endo et al., 2008). Primers used were Q2-ANAC089 and 18S rRNA as a standard control (Table S3).
RNA extraction for microarray analysis
Total RNA was extracted from 3-day-stratified seeds and 3 hours untreated or 3 μM ABA treated Col-0 and gap1-2 seeds under light conditions as previously described (Oñate-Sánchez and Vicente-Carbajosa, 2008).
Synthesis of biotinylated cRNA for microarrays analysis
Each RNA preparation was tested for degradation using the Agilent 2100 Bioanalyzer (Agilent technologies, Palo Alto, CA). cDNA was synthesized from 4 μg of total RNA using One-cycle target labeling and control reagents (Affymetrix, Santa Clara, CA) to produce biotin labeled cRNA. The cRNA preparations (15 μg) were fragmented at 94°C for 35 min into 35-200 bases in length.
Hybridization, washing, and scanning of microarrays
Three biological replicates for each condition were independently hybridized. If the quality control was correct, then 10 μg of fragmented cRNA were hybridized to the Arabidopsis ATH1 Genome array (Affymetrix, Santa Clara, CA), containing 22500 transcript variants from 24000 well -characterized Arabidopsis thaliana genes. Each sample was added to a hybridization solution containing 100mM 2-(N-morpholino) ethanesulfonic acid, 1 M Na+, and 20mM of EDTA in the presence of 0.01% of Tween-20 to a final cRNA concentration of 0.05 μg/ml. Hybridization was performed for 16 h at 45°C. Each microarray was washed and stained with streptavidin-phycoerythrin in a Fluidics station 450 (Affymetrix) and scanned at 2.5 μm resolution in a GeneChip® Scanner 3000 7G System (Affymetrix) Data analysis were performed using GeneChip Operating Software (GCOS).
Data analysis for microarrays experiments
Analysis was performed using the affylmaGUI R package (Wettenhall et al., 2006). Robust Multi-array Analysis (RMA) algorithm was used for background correction, normalization, and expression levels summarization (Irizarry et al., 2003). Next, differential expression analysis was performed with the Bayes t-statistics from the linear models for Microarray data (limma), included in the affylmGUI package. P values were corrected for multiple-testing using the Benjamini-Hochberg’s method (False Discovery Rate) (Benjamini and Hochberg, 1995; Reiner et al., 2003). Genes were considered to be differentially expressed if the corrected P values were < 0.05. In addition, only genes with a signal log ratio > 1 or < −1 were considered for further analysis.
Corregulation analysis, functional categories, and gene classification
Corregulation studies were performed between gap1-2 and different germination conditions. Two different expression categories were considered, one corresponding to all genes induced (≥2-fold) or repressed (≤-2-fold) in both conditions compared, and other corresponding to genes showing high induction (≥2-fold) or repression (≤-2-fold) in one condition and in the other condition at least minimum induction (1-fold ≤ x,y < 2-fold) or repression (−1-fold ≥ x,y > −2-fold), respectively. A total of 465 genes were analyzed. The hierarchical cluster was calculated and drawn using The Institute for Genomic Research (TIGR) MeV (Multiarray experiment Viewer, version 4.4) software provided by the TIGR Institute (Saeed et al., 2003) and using Pearson uncentered and complete linkage. CEL.files for the different treatments were obtained from AtGenExpress project (http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp), and all the selected datasets were normalized using RMAExpress and analyzed. The display and gene classifications are based on MapMan (Thimm et al., 2004; Usadel et al., 2005). A full description of the bins and layout can be obtained at http://www.gabi.rzpd.de/projects/MapMan/, version 3.0.0.
Universal protein binding microarray 11 (PBM11) for determining TFs DNA-binding specificities
Godoy et al. (2011) designed a DNA microarray containing all possible DNA sequences of 11 nucleotides (4,194,325 sequences) compacted in 167,773 35 nucleotides-long oligonucleotides, so each probe contained 25 overlapping 11-nt sequences. The design was based on the properties of de Bruijn sequence of order 11 with an alphabet of size 4 (ATCG; Berger et al., 2006; Philippakis et al., 2008). To determine the sequence of every oligonucleotide probe in the microarray the de Bruijn sequence was divided into 167,773 35-nt fragments overlapping 10 nucleotides to ensure that all subsequences of size 11 were included. All the probes started with the common 25-nt sequence (5′-TGTCTGTGTTCCGTTGTCCGTGCTG-3′). Microarrays of 267x912 single-stranded 60-nt oligonucleotides were manufactured by Agilent Technologies. The difference between available spots and 167,773 different probes was occupied with 73,626 probes randomly chosen, in a way that 94,147 probes were represented once in the PBM11 and 73,626 twice.
Synthesis of double-stranded microarray, protein incubation, and immunological detection of DNA-protein complexes for the PBM11
Synthesis in situ of double-stranded DNA and processing of slides were as in Berger and Bulyk (2009) but omitting the blocking steps. The binding mixture was adjusted to 100 μl and contained 2% milk, 100 ng of poly deoxyinosinic-deoxycytidylic acid (dI-dC), 0.5 μg of denatured salmon sperm DNA (ssDNA), 1 μg of purified protein and 25 μl of binding buffer 4x (10mM Tris-HCl pH 8, 60mM KCl, 4mM MgCl2, 0,1mM EDTA, 10% glycerol, 200mg/ml BSA). For incubation with ANAC089 recombinant protein expressed in planta, the binding mixture contained dI-dC, ssDNA and milk as before, and 70 μl of protein extract isolated from N. benthamiana leaves (Godoy et al., 2011). The binding mixture was deposited onto the slide, covered with a LifterSlip (22x65 mm; Erie Scientific) and incubated in a humid chamber for 2.5 hours at room temperature. Slides were washed 3 times in 50 mL of Phosphate Buffered Saline (PBS)-1% Tween 20 (5 minutes), 3x in PBS-0.01% Triton X-100 (5 minutes) and spun dry by centrifugation. DNA-protein complexes were incubated with 16 μg of Rabbit polyclonal to Maltose Binding Protein (Abcam) in PBS-2% milk for 16 h at room temperature. Slides were washed 3x in PBS- 0.05% Tween 20, 3x in PBS-0.01% Triton X-100 (5 minutes each wash) and dried. Labeling of DNA-protein complexes were performed by incubating the microarrays with 0.4 μg of goat anti-rabbit IgG DyLight 549 conjugated (Pierce) in PBS-2% milk for 3 hours at room temperature, followed by the same washes as before and the slides dried for scanning.
Image acquisition, data processing, and analysis of PBM11
Two different images were obtained for each microarray at 5 μm in a GenePix 4000B scanner (Axon Instruments) and quantified in the GenePix Pro 5.1 software. One corresponded to double-stranded DNA at 635 nm, generating “control DNA” GPR files. The second image was obtained after labeling of DNA-protein complexes at 532 nm, generating “binding” GPR files. Combining of “control” and “binding” files, normalization and adjustment of the probe intensities and transformation to a list of scores for all the k-mers considered were carried out with the PBM Analysis Suite from Berger and Bulyk (2009); http://the_brain.bwh.harvard.edu/PBMAnalysisSuite/index.html. We adapted these scripts to the dimensions of our PBM11 microarray and for calculating average intensities of duplicated probes. The best scored motif, represented as an energy- based matrix, was converted into a graphical logo with the on-line tool enoLOGOS (http://chianti.ucsd.edu/cgi-bin/enologos/enologos.cgi).
Production of recombinant ANAC089 protein and polyclonal antibodies
ANAC089 cDNA was recombined into pET-28a+ vector to obtain fusion protein with N-terminal His-tag. Recombinant protein was expressed in Escherichia coli and purified using His-Select® Nickel Affinity Gel (IMAC) (Sigma) according to manufacturer (Biomedal). A primary dose of 750 μg purified recombinant protein 6xHis-ANAC089 was emulsified in Freunds complete adjuvant (Sigma) and administered subcutaneously in two rabbits. Three doses of the protein (375 μg) emulsified in Freunds incomplete adjuvant (Sigma) were administered at intervals (21 days). After the third booster injection (10 days), blood was collected from the rabbits and the serums were separated. Antibodies (Anti-ANAC089) were isolated by column chromatography with a protein-G column (GE Healthcare Life Sciences).
Western blot
Total proteins from of Col-0, Ler, gap1-1, gap1-2, gap1-3, 35S:GFP-ANAC089 and pANAC089:ANAC089ΔC-1-GFP lines were extracted after the indicated treatments and times for western blot analysis. Tissue was powdered with mortar and pestle and incubated for 10 min on ice with extraction buffer (50mM Tris-HCl 7.5 pH, 75mM NaCl, 15mM EGTA, 15mM MgCl2, 1mM DTT, 0.1% Tween 20, 1mM NaF, 0.2M NaV, 2mM Na-pyrophosphate, 60mM β-glycerolphosphate and 1x proteases inhibitor mix, Roche) followed by centrifugation for 10 min at 13000 rpm at 4°C. Protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad) based on the Bradford method (Bradford, 1976). 60 μg of total protein was loaded per well in SDS-acrylamide/bisacrylamide gel electrophoresis using Tris-glycine-SDS buffer. Proteins were electrophoretically transferred to an InmobilonTM-P PVDF membrane (Millipore) using the Trans-Blot® Turbo (Bio-Rad). Membranes were blocked in TBS-T containing 5% Blocking Agent and probed with antibodies diluted in blocking buffer. Anti-ANAC089 Purified Rabbit Immunoglobulin (Biomedal), anti-ABI5 Purified Rabbit Immunoglobulin (Biomedal), Anti-GFP (Clontech), Anti-Actin clone 10-B3 Purified Mouse Immunoglobulin (Sigma) and ECL-Peroxidase labeled anti-rabbit and anti-mouse (Amersham) antibodies were used in the western blot analyses. Detection was performed using ECL Advance Western Blotting Detection Kit (Amersham) and the chemiluminescence was detected using an Intelligent Dark-Box II, LAS-1000 scanning system (Fujifilm). Quantification of band intensity was performed with ImageJ software.
Fluorescence microscopy
7-day-old 35S:GFP-ANAC089 plants were treated during 4 hours with 1 mM cPTIO, 1 mM DTT, 500 μM GSNO, 1.5 mM GSH, 1.5 mM GSSG, 1 μM ABA or without treatment (control) for changes in subcellular localization of ANAC089. The fluorescent photographs were taken using a Leica SP5 confocal microscope and Bio-Rad Radiance 2100 laser scanning confocal imaging system (LaserSharp v.5 Image software). For GFP detection, the excitation source was an argon ion laser at 488 nm and detection filters between 481 and 530 nm.
Nuclear/cytoplasmic fractioning assays
7-day-old 35S:GFP-ANAC089 plants were treated during 2 or 4 hours with 1 mM cPTIO or 1 mM DTT and the corresponding control and immediately harvested to proceed with subcellular fractioning studies. Briefly, 1,5 g of sample were mortar and pestle-grounded and resuspended in 5 mL of 1X NIB buffer (CelLytic™PN, Sigma) supplemented with freshly prepared 1 mM DTT. The suspension was filtered and centrifuged at 1,260 g for 10 min at 4°C. Pellets were resuspended in 1 mL of NIBA (1X NIB, 1X cOmplete® EDTA-free proteases inhibitor) 0.3% Triton X-100 and centrifuged at 1,200 g for 10 min at 4°C. The supernatants were collected as the cytosolic fraction whereas pellets were washed five times in 1 mL of NIBA for crude nuclei preparation. Protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad) based on the Bradford method (Bradford, 1976). 20 μg of total protein per sample was loaded to proceed with western blot experiments. Anti-ANAC089 Purified Rabbit Immunoglobulin (Biomedal) to detect ANA089, and anti-H3 histone (Agrisera) and anti-RuBisCo-Large subunit (Agrisera) were used to determine the pureness of nuclear and cytoplasmic subcellular fractions, respectively.
Quantification and statistical analysis
Information about statistical tests used and technical and biological replicate experiments for each experiment are detailed in the relevant STAR Methods sections. Values are represented as mean ± SE. The statistical analysis were performed by a two tailed Student’s t test where p < 0.05, p < 0.01, p < 0.001 are considered significant and expressed as ∗, ∗∗, ∗∗∗, respectively.
Acknowledgments
We thank the Spanish networks BIO2015-68957-REDT and RED2018-102397-T for stimulating discussions, as well as Dr. José M. Carrasco and Dr. Pablo Vera (IBMCP-CSIC) for help with the protein-expression experiments of the PBM. This work was financed by grants EcoSeed Impacts of Environmental Conditions on Seed Quality “EcoSeed-311840” ERC.KBBE.2012.1.1-01; BIO2017-85758-R and CSD2007-00057 (TRANSPLANTA) from the Ministerio de Ciencia, Innovación y Universidades (MICIU) (Spain); SA313P18 and SA137P20 from Junta de Castilla y León; Escalera de Excelencia CLU-2018-04 co-funded by the P.O. FEDER of Castilla y León 2014–2020 Spain (to O.L.); and the PhD and University Teacher Training Fellowship, Spanish Ministry of Science and Education (to P.A.).
Author contributions
P.A. performed most of the experiments, analyzed the data, and wrote the manuscript. K.T., K.N., and E.N. performed the genetic screening, mapping, and cloning, analyzed the data, and provided essential reagents. I.M., I.S.-V., and A.F.-A. performed the germination, the subcellular fractioning, the ROS detection, and the transcriptomic assays. M.G., J.M.F., and R.S. performed the microarray and contributed to the experimental design of PBM11 and the statistical analysis. D.G., T.R., W.S., and I.K. measured the ABA, thiol, and disulfide levels and analyzed the data. C.P.-R. and J.S. performed the cold stress assays and contributed to the interpretation. O.L. conceived the study, designed the experiments, and wrote the manuscript. All of the authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: June 15, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109263.
Supplemental information
Position of the cis-element binding site of ANAC089 within 1 kb of promoter region of these genes (p value < 0.01). Highlighted in pink: Redox related (ascorbate; glutathione; hypoxia; oxidative stress); in blue: Cysteine-type endopeptidases and serine-type carboxypeptidases; in green: hormone metabolism. Genes highlighted in bold with the specific cis-acting elements recognized by the ANAC089 TF.
Position of the cis-element binding site of ANAC089 within 1 kb of the promoter region of these genes (p value < 0.01). Highlighted in pink: Redox related (ascorbate; glutathione; hypoxia; oxidative stress); in yellow: dormancy related genes; in green: hormone metabolism. Genes highlighted in bold with the specific cis-acting elements recognized by the ANAC089 TF.
References
- Albertos P., Romero-Puertas M.C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., Lorenzo O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015;6:8669. doi: 10.1038/ncomms9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C., Kranner I. Analyses of reactive oxygen species and antioxidants in relation to seed longevity and germination. Methods Mol. Biol. 2011;773:343–367. doi: 10.1007/978-1-61779-231-1_20. [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.-I., Köhler B., Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62:250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- Beligni M.V., Lamattina L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta. 2000;210:215–221. doi: 10.1007/PL00008128. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to MultipleTesting. J. R. Stat. Soc. Ser. A Stat. Soc. 1995;57:289–300. [Google Scholar]
- Berger M.F., Bulyk M.L. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., 3rd, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P.C., Badger M.R., Jones R.L. Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell. 2004;16:332–341. doi: 10.1105/tpc.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P.C., Gubler F., Jacobsen J.V., Jones R.L. Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta. 2004;219:847–855. doi: 10.1007/s00425-004-1282-x. [DOI] [PubMed] [Google Scholar]
- Bewley J.D., Black M. Seeds Physiology of Development and Germination. J. Ecol. 1994;83:1053. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Castillo M.C., Lozano-Juste J., González-Guzmán M., Rodriguez L., Rodriguez P.L., León J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015;8:ra89. doi: 10.1126/scisignal.aaa7981. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Costa-Broseta Á., Perea-Resa C., Castillo M.-C., Ruíz M.F., Salinas J., León J. Nitric Oxide Controls Constitutive Freezing Tolerance in Arabidopsis by Attenuating the Levels of Osmoprotectants, Stress-Related Hormones and Anthocyanins. Sci. Rep. 2018;8:9268. doi: 10.1038/s41598-018-27668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Broseta Á., Perea-Resa C., Castillo M.C., Ruíz M.F., Salinas J., León J. Nitric oxide deficiency decreases C-repeat binding factor-dependent and -independent induction of cold acclimation. J. Exp. Bot. 2019;70:3283–3296. doi: 10.1093/jxb/erz115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic. Acids Res. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J., Neill S. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2002;99:16314–16318. doi: 10.1073/pnas.252461999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Sawada Y., Takahashi H., Okamoto M., Ikegami K., Koiwai H., Seo M., Toyomasu T., Mitsuhashi W., Shinozaki K. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008;147:1984–1993. doi: 10.1104/pp.108.116632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M., Sanz L., Lewis D.R., Muday G.K., Lorenzo O. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. USA. 2011;108:18506–18511. doi: 10.1073/pnas.1108644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., Sechet J., To A., North H.M., Marion-Poll A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012;70:501–512. doi: 10.1111/j.1365-313X.2011.04887.x. [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Gibbs D.J., Md Isa N., Movahedi M., Lozano-Juste J., Mendiondo G.M., Berckhan S., Marín-de la Rosa N., Vicente Conde J., Sousa Correia C., Pearce S.P. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell. 2014;53:369–379. doi: 10.1016/j.molcel.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M., Franco-Zorrilla J.M., Pérez-Pérez J., Oliveros J.C., Lorenzo O., Solano R. Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 2011;66:700–711. doi: 10.1111/j.1365-313X.2011.04519.x. [DOI] [PubMed] [Google Scholar]
- He X.J., Mu R.L., Cao W.H., Zhang Z.G., Zhang J.S., Chen S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- Jiang H., Li H., Bu Q., Li C. The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signal. Behav. 2009;4:464–466. doi: 10.4161/psb.4.5.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Jikumaru Y., Hanada A., Nambara E., Abrams S.R., Kamiya Y., Seo M. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010;51:1988–2001. doi: 10.1093/pcp/pcq158. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Kim S.G., Kim Y.S., Seo P.J., Bae M., Yoon H.K., Park C.M. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35:203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G., Lee A.K., Yoon H.K., Park C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- Klein P., Seidel T., Stöcker B., Dietz K.J. The membrane-tethered transcription factor ANAC089 serves as redox-dependent suppressor of stromal ascorbate peroxidase gene expression. Front. Plant Sci. 2012;3:247. doi: 10.3389/fpls.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang J., Wang X., Chen J. A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana. Sci. China Life Sci. 2010;53:1299–1306. doi: 10.1007/s11427-010-4085-2. [DOI] [PubMed] [Google Scholar]
- Li P., Wind J.J., Shi X., Zhang H., Hanson J., Smeekens S.C., Teng S. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc. Natl. Acad. Sci. USA. 2011;108:3436–3441. doi: 10.1073/pnas.1018665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhou H., Shi X., Yu B., Zhou Y., Chen S., Wang Y., Peng Y., Meyer R.C., Smeekens S.C., Teng S. The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 2014;10:e1004213. doi: 10.1371/journal.pgen.1004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Durner J. Interplay of reactive oxygen species and nitric oxide: nitric oxide coordinates reactive oxygen species homeostasis. Plant Physiol. 2015;167:1209–1210. doi: 10.1104/pp.15.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shi L., Ye N., Liu R., Jia W., Zhang J. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009;183:1030–1042. doi: 10.1111/j.1469-8137.2009.02899.x. [DOI] [PubMed] [Google Scholar]
- Malik S.I., Hussain A., Yun B.W., Spoel S.H., Loake G.J. GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci. 2011;181:540–544. doi: 10.1016/j.plantsci.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Mao X., Zhang H., Qian X., Li A., Zhao G., Jing R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012;63:2933–2946. doi: 10.1093/jxb/err462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Mandon J., Cristescu S.M., Harren F.J.M., Prats E. Methods of nitric oxide detection in plants: a commentary. Plant Sci. 2011;181:509–519. doi: 10.1016/j.plantsci.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Mur L.A.J., Mandon J., Persijn S., Cristescu S.M., Moshkov I.E., Novikova G.V., Hall M.A., Harren F.J.M., Hebelstrup K.H., Gupta K.J. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5:pls052. doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. A Revised Medium for Rapid Growth and Biol. Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Nakabayashi K., Bartsch M., Xiang Y., Miatton E., Pellengahr S., Yano R., Seo M., Soppe W.J.J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell. 2012;24:2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013;32:959–970. doi: 10.1007/s00299-013-1418-1. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Nambara E., Suzuki M., Abrams S., McCarty D.R., Kamiya Y., McCourt P. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics. 2002;161:1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A.M., Satoh K., Kondoh H., Ooka H., Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kumar A., Li W., Wang Y., Siddiqi M.Y., Crawford N.M., Glass A.D.M. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006;140:1036–1046. doi: 10.1104/pp.105.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Lo Leggio L., Johansson E., Larsen S., Skriver K. Preliminary crystallographic analysis of the NAC domain of ANAC, a member of the plant-specific NAC transcription factor family. Acta Crystallogr. D Biol. Crystallogr. 2004;60:112–115. doi: 10.1107/s0907444903022029. [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., Matsubara K., Osato N., Kawai J., Carninci P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Park J., Kim Y.S., Kim S.G., Jung J.H., Woo J.C., Park C.M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011;156:537–549. doi: 10.1104/pp.111.177071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis A.A., Qureshi A.M., Berger M.F., Bulyk M.L. Design of compact, universal DNA microarrays for protein binding microarray experiments. J. Comput. Biol. 2008;15:655–665. doi: 10.1089/cmb.2007.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.L., Ward D.A., Uknes S., Appleford N.E., Lange T., Huttly A.K., Gaskin P., Graebe J.E., Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchet E., Verdu I., Delahaie J., Cukier C., Girard C., Morère-Le Paven M.-C., Limami A.M. Abscisic acid-induced nitric oxide and proline accumulation in independent pathways under water-deficit stress during seedling establishment in Medicago truncatula. J. Exp. Bot. 2014;65:2161–2170. doi: 10.1093/jxb/eru088. [DOI] [PubMed] [Google Scholar]
- Preston J., Tatematsu K., Kanno Y., Hobo T., Kimura M., Jikumaru Y., Yano R., Kamiya Y., Nambara E. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 2009;50:1786–1800. doi: 10.1093/pcp/pcp121. [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Saavedra X., Modrego A., Rodríguez D., González-García M.P., Sanz L., Nicolás G., Lorenzo O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010;152:133–150. doi: 10.1104/pp.109.146381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sánchez-Montesino R., Bouza-Morcillo L., Marquez J., Ghita M., Duran-Nebreda S., Gómez L., Holdsworth M.J., Bassel G., Oñate-Sánchez L. A Regulatory Module Controlling GA-Mediated Endosperm Cell Expansion Is Critical for Seed Germination in Arabidopsis. Mol. Plant. 2019;12:71–85. doi: 10.1016/j.molp.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vicente I., Fernández-Espinosa M.G., Lorenzo O. Nitric oxide molecular targets: reprogramming plant development upon stress. J. Exp. Bot. 2019;70:4441–4460. doi: 10.1093/jxb/erz339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Fernández-Marcos M., Modrego A., Lewis D.R., Muday G.K., Pollmann S., Dueñas M., Santos-Buelga C., Lorenzo O. Nitric oxide plays a role in stem cell niche homeostasis through its interaction with auxin. Plant Physiol. 2014;166:1972–1984. doi: 10.1104/pp.114.247445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Albertos P., Mateos I., Sánchez-Vicente I., Lechón T., Fernández-Marcos M., Lorenzo O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015;66:2857–2868. doi: 10.1093/jxb/erv213. [DOI] [PubMed] [Google Scholar]
- Schausberger C., Roach T., Stöggl W., Arc E., Finch-Savage W.E., Kranner I. Abscisic acid-determined seed vigour differences do not influence redox regulation during ageing. Biochem. J. 2019;476:965–974. doi: 10.1042/BCJ20180903. [DOI] [PubMed] [Google Scholar]
- Schneider C., Rasband W., Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Park J.Y., Kim S.Y., Jeon J., Lee Y.H., Kim J., Park C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- Smyczynski C., Roudier F., Gissot L., Vaillant E., Grandjean O., Morin H., Masson T., Bellec Y., Geelen D., Faure J.D. The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J. Biol. Chem. 2006;281:25475–25484. doi: 10.1074/jbc.M601815200. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Phillips A.L., Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O.E., Palacios-Rojas N., Selbig J., Hannemann J., Piques M.C., Steinhauser D. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Hou Y.J., Zhao Y., Hsu C.C., Yuan F., Zhu X., Tao W.A., Song C.P., Zhu J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA. 2015;112:613–618. doi: 10.1073/pnas.1423481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhu J.K., Lang Z. Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal. Behav. 2015;10:e1031939. doi: 10.1080/15592324.2015.1031939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J.M., Simpson K.M., Satterley K., Smyth G.K. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- Yang Z.T., Wang M.J., Sun L., Lu S.J., Bi D.L., Sun L., Song Z.T., Zhang S.S., Zhou S.F., Liu J.X. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014;10:e1004243. doi: 10.1371/journal.pgen.1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.G., Chen L., Zhang L.L., Zhang W.H. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol. 2009;151:755–767. doi: 10.1104/pp.109.140996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Cai S., Wang Y., Chen Z.H. Loss of nitrate reductases NIA1 and NIA2 impairs stomatal closure by altering genes of core ABA signaling components in Arabidopsis. Plant Signal. Behav. 2016;11:e1183088. doi: 10.1080/15592324.2016.1183088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Position of the cis-element binding site of ANAC089 within 1 kb of promoter region of these genes (p value < 0.01). Highlighted in pink: Redox related (ascorbate; glutathione; hypoxia; oxidative stress); in blue: Cysteine-type endopeptidases and serine-type carboxypeptidases; in green: hormone metabolism. Genes highlighted in bold with the specific cis-acting elements recognized by the ANAC089 TF.
Position of the cis-element binding site of ANAC089 within 1 kb of the promoter region of these genes (p value < 0.01). Highlighted in pink: Redox related (ascorbate; glutathione; hypoxia; oxidative stress); in yellow: dormancy related genes; in green: hormone metabolism. Genes highlighted in bold with the specific cis-acting elements recognized by the ANAC089 TF.
Data Availability Statement
The published article includes all raw and analyzed datasets generated during this study. Original data have been deposited to NCBI GEO: GSE25159. Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession number: NAC089, AT5G22290.