Figure 2.
Integration of BioID and PRISMA data revealed a spatial CEBPA interactome with high interconnectivity
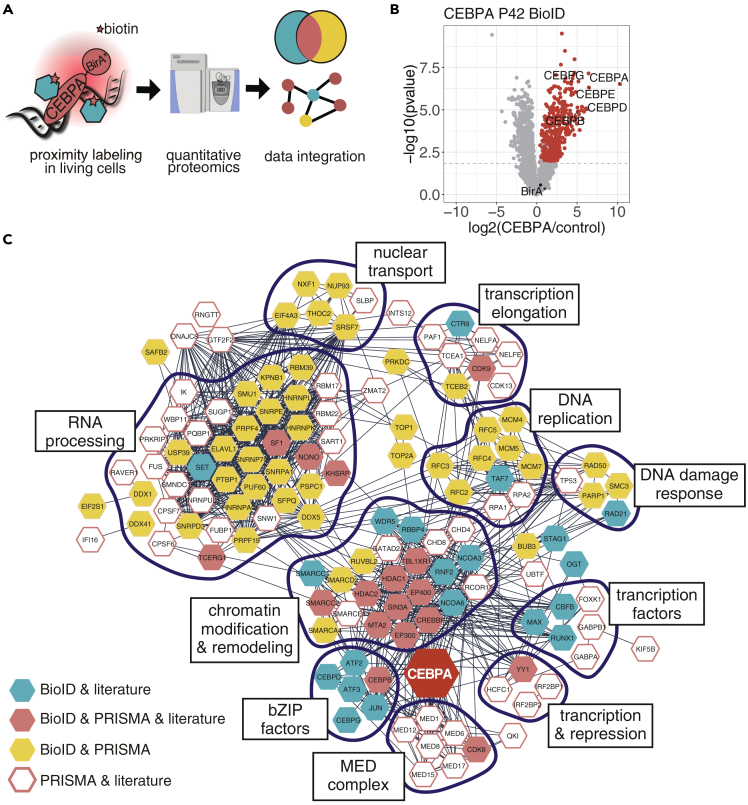
(A) BioID proximity labeling data obtained in living cells was integrated with biochemical data obtained with PRISMA.
(B) The CEBPA BioID interactome in NB4 cells. Log fold changes (CEBPA/control) of proteins are plotted against their p-values (student t-test, n = 4). The FDR cutoff (0.05) is indicated with a dotted line.
(C) Integrated protein interaction network of CEBPA. Color of nodes corresponds to detection in datasets as depicted in the legend. Edges represent validated protein interactions retrieved from the STRING database. Interactors not connected by any edges were removed from the plot (12 proteins).
See also Figures S3 and S4, Table S4.