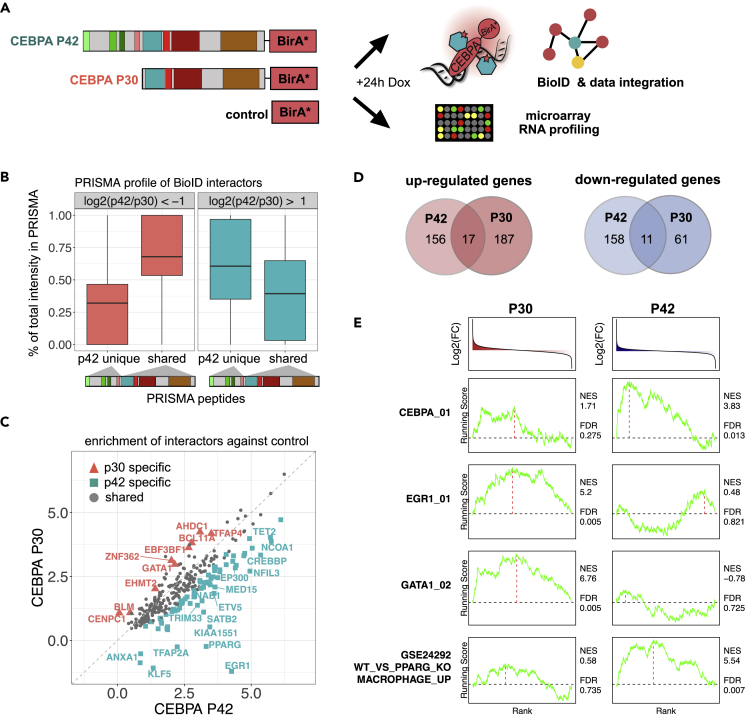
Figure 4.
BioID with CEBPA p42/p30 isoforms disclose isoform-specific interactors in conjunction with PRISMA profiles
(A) BioID interaction proteomics (n = 4) and microarray RNA expression (n = 3) were performed with CEBPA isoform (p42 or p30) expressing cells.
(B) PRISMA profiles of interactors showing preference for p42 (log2(p42/p30) > 1) or p30 (log2(p42/p30) < - 1) in BioID. LFQ intensities of proteins were summed across PRISMA peptides corresponding to exclusively p42 or both isoforms. The resulting numbers were divided by the total LFQ intensity of each protein across all PRISMA peptides. PTM-modified peptides were omitted from the calculations.
(C) Relative enrichment of CEBPA interactors against the control, p42 is plotted against p30. Proteins marked in color passed 0.1 FDR threshold in a direct comparison.
(D) Overlap of up- and downregulated genes as detected by microarray gene expression profiling (comparison to BirA∗ cells, FDR <0.05, absolute fold change (FC) > 1).
(E) Induced gene expression changes were subjected to ssGSEA analysis. Normalized enrichment score (NES) and FDR of informative gene sets are displayed next to running score line plots. The running score was calculated with Kolmogorov-Smirnov (K-S) running sum statistics.