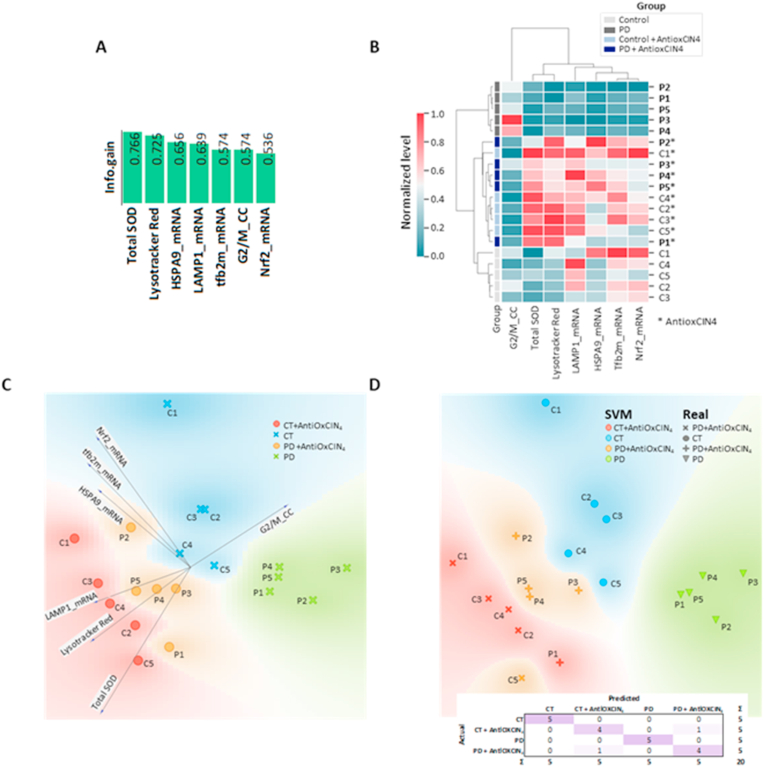
Fig. 12.
Integrative data analysis. A) Mutual Information (Information Gain) evidenced total SOD, Lysotracker Red staining, HSPA9, LAMP1 and TFB2m transcripts, as well as G2/M phase (cell cycle) and NRF2 transcripts as the features with more discriminative power between all experimental conditions. B) Selection of the 7 most discriminant features allowed a perfect separation of untreated control and PD samples into distinct clusters, while samples treated with AntiOxCIN4 were placed in the same cluster. C) PCA analysis using the same 7 features evidenced an excellent separation between the experimental conditions. D) Supervised machine learning analysis using support vector machines with cross-validation provided a Precision and Recall of 0.900 each, with an area under ROC curve of 0.887 and Classification Accuracy and F1 score of 0.900 each. Only two samples were misclassified (C5 + AntiOxCIN4 classified as PD + AntiOxCIN4 and PD1 + AntiOxCIN4 classified as C + AntiOxCIN4). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)