Figure 1. Schematic Representation of the Various Analyses Performed in the Study.
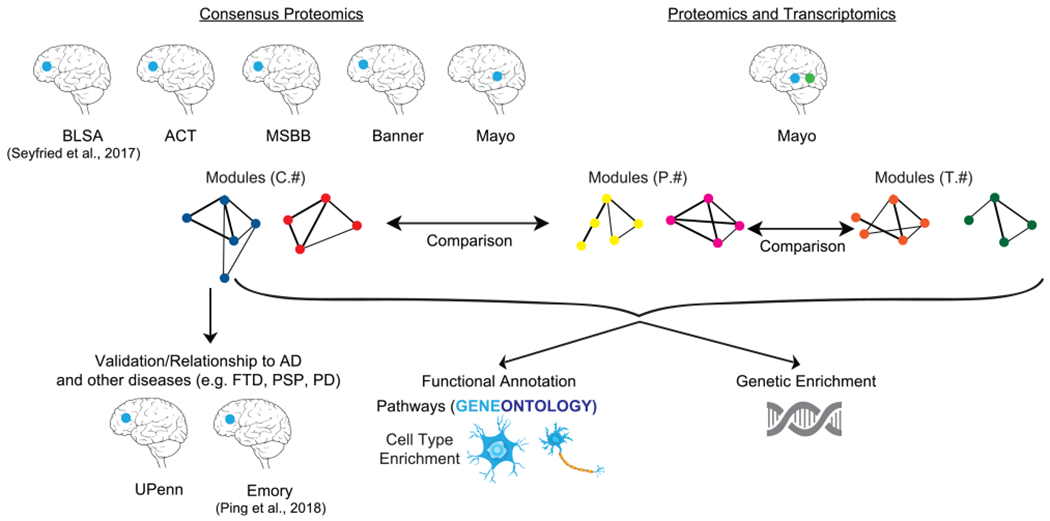
Consensus proteomics modules were generated from five different studies: the Baltimore Longitudinal Study of Aging (BLSA), Adult Changes of Thought (ACT), Mount Sinai Brain Bank (MSBB), Banner Sun Health Research Institute (Banner), and Mayo Clinic Brain Bank (Mayo). Furthermore, the Mayo dataset had matched samples where proteomics and transcriptomics data were available. Comparisons were then made among the consensus proteomics modules (labeled with “C”), Mayo-only proteomics modules (labeled with “P”), and Mayo-matched sample transcriptomics modules (labeled with “T”). Determining the relationship of consensus proteomic modules with other neurodegenerative diseases like frontotemporal dementia (FTD), progressive supranuclear palsy (PSP), and Parkinson’s disease (PD) was performed using completely separate datasets (UPenn and Emory). Functional annotation of the modules was performed using cell type and Gene Ontology. Common genetic enrichment analysis was used to understand the relationship of modules with causal genetic drivers. The blue dot indicates the location of proteomic brain samples. The green dot indicates location of transcriptomic brain samples.
