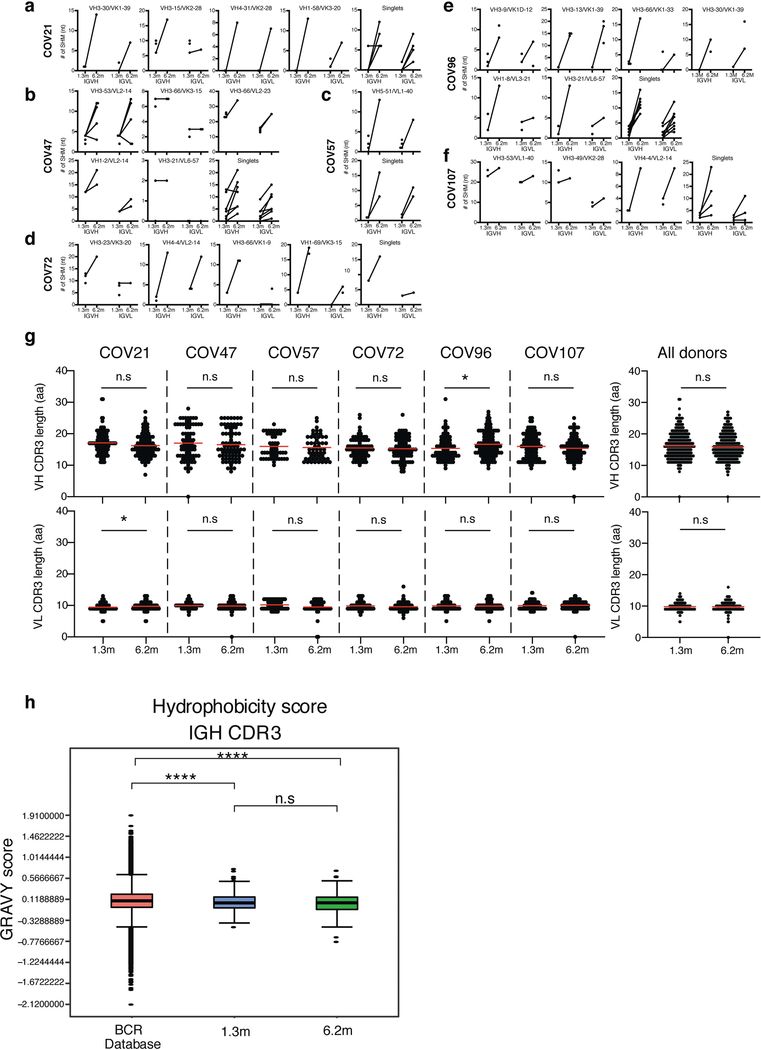
Extended Data Fig. 6 |. Analysis of antibody somatic hypermutation of persisting clones, CDR3 length and hydrophobicity.
a–f, Number of somatic nucleotide mutations in both the IGVH and IGVL of persisting clones found at month-1.3 and month-6.2 time points in six participants: COV21 (a), COV47 (b), COV57 (c), COV72 (d), COV96 (e) and COV107 (f). The VH and VL gene usage of each clonal expansion is indicated above the graphs, or are indicated as ‘singlets’ if the persisting sequence was isolated only once at both time points. Connecting line indicates the somatic hypermutation of the clonal pairs that were expressed as a recombinant monoclonal antibodies. g, For each individual, the amino acid length of the CDR3 at IGVH and IGVL is shown. The horizontal bars indicate the mean. The number of antibody sequences (IGVH and IGVL) evaluated for each participant are n = 90 (COV21), n = 78 (COV47), n = 53 (COV57), n = 87 (COV72), n = 104 (COV96), n = 120 (COV107). Right, all antibodies combined (n = 532 for both IGVH and IGVL). Statistical significance was determined using two-tailed Mann–Whitney U-tests and horizontal bars indicate median values. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. h, Distribution of the hydrophobicity GRAVY scores at the IGH CDR3 in 532 antibody sequences from this study compared to a public database: statistical analysis is provided in Methods. The box limits are at the lower and upper quartiles, the centre line indicates the median, the whiskers are 1.5× interquartile range and the dots represent outliers. Statistical significance was determined using two-tailed Wilcoxon matched-pairs signed-rank test. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).