In efforts to identify cell type-specific causal processes in vascular diseases, our research communities have rapidly evolved from use of a single or a few pre-selected markers to non-biased tissue and single-cell analyses. Such analyses of human and mouse tissues have provided insights into the amazing diversity of cell populations in vascular diseases and atherosclerotic lesions.1 Non-biased “omics” approaches, including transcriptomics, proteomics and metabolomics, are now increasingly used in combination with single-cell technologies and spatial profiling to investigate vascular diseases.
A general model for the discovery and validation of novel pathways and drug targets using integration of non-biased -omics and single-cell approaches is described in Figure 1. The discovery phase compares human diseased tissues with healthy control tissues and tissues from animal models of the disease. Because differences in cell type composition of diseased and normal tissues can introduce bias when using bulk tissue profiling, singe-cell non-biased -omic strategies are most informative. The integrative computational phase makes use of experimental data and publicly available resources on human disease, cross-species data, and cross-platform data. For example, cross-species reference-based analyses of single cell data2, 3 give a framework for integration of experimental and genetic manipulations in model systems to provide greater mechanistic insights into human disease. Similarly, cross-platform integration of public human genetic data facilitates causal and directional inference for genes, proteins and metabolites in a cell-specific context.4 The molecular functions of novel targets and pathways as well as computationally predicted master regulators3, 6 can be investigated in isolated cells and using gene-editing technologies and human induced pluripotent stem cell systems. Validation is critical to prove causality and significance of identified pathways and specific targets, first in animal models and then, through clinical translation, in cogent clinical trials. This strategy can ultimately leverage precision medicine principles and targeted therapeutics, which allow treatment of patients according to their specific set of risk factors and molecular and cellular disruptions.
Figure 1. Model for discovery and validation of novel drug targets for complex diseases using integrative single-cell non-biased -omics.
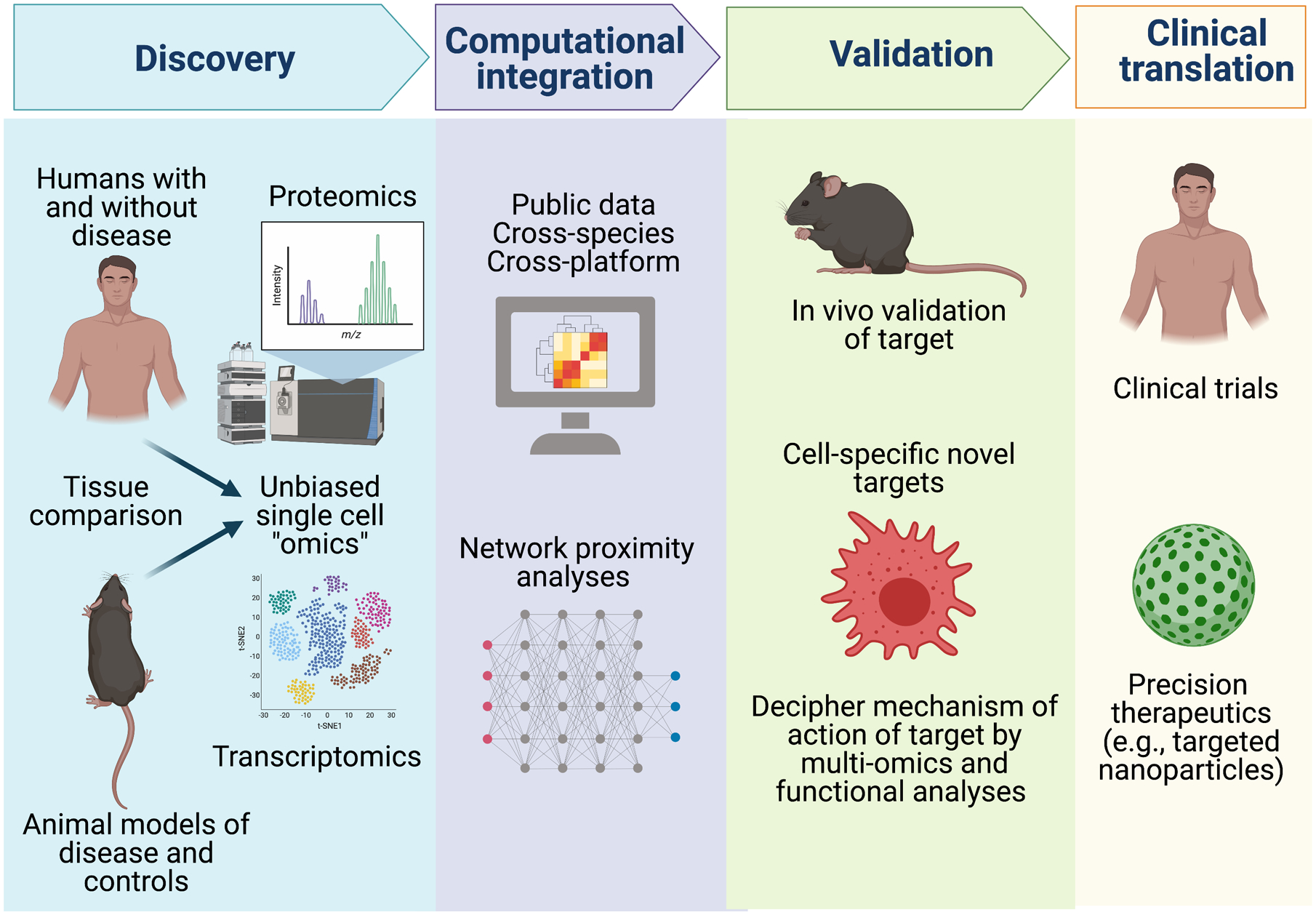
A discovery phase using both human disease and animal model single cell -omics profiling is recommended. Integrative computational methods are applied to cross-species and cross-platform data to identify and prioritize master regulatory and druggable targets of greatest importance in human disease. Experimental functional studies in animal models of disease and in disease-relevant human cells, including gene-manipulation in human induced pluripotent stems cells, can define precise molecular and cellular targets for translation via clinical trials and precision therapeutic strategies. Figure created with BioRender.com.
Using elements of the approach in Figure 1, in this issue of Circulation Decano and colleagues describe a proteomics-driven systems approach to identify drug target candidates for the treatment of vein graft failure5. Vein grafts are used to treat peripheral artery disease and as coronary artery bypass grafts. However, many of these grafts become occluded and fail with time, developing lesions in part similar to those in atherosclerotic arteries. There are currently no effective treatments available for prevention of vein graft failure, as statins and other drugs that reduce atherosclerotic cardiovascular disease (CVD), do not appear to be effective.
Decano et al.5 reasoned that a non-biased approach would be most useful for identification of potential novel drug targets. In a series of elegant mouse studies, the authors first utilized global tissue proteomics to analyze the inferior vena cava from donor mice sutured to the carotid artery to create vein grafts in a small number of fat-fed LDL receptor-deficient (Ldlr−/−) recipient mice and wildtype mice fed a standard rodent diet (discovery phase, Figure 1). Vein grafts in fat-fed Ldlr−/− mice developed lesions containing smooth muscle cells and macrophages and had a proteome composition markedly different from that of the control undisturbed vena cava. Proteomics performed at several different time-points up to 4 weeks after vein grafting identified 30 protein clusters regulated in temporally distinct fashions in vein grafts versus the undisturbed vena cava. Because single-cell proteomics was not used in the discovery phase, a significant portion of the identified changes in tissue proteome was likely due to differences in tissue cell type composition. Caution is also warranted given the lack of primary non-biased discovery in human diseased tissues, particularly in considering the relevance of discoveries in a severely hyperlipidemic mouse model to the human graft disease, which often emerges despite low circulating lipoproteins and aggressive treatment of hypercholesterolemia.
Pathway networks were then constructed, and the most central proteins in each pathway were computed. An overall network associated with inflammation, response to toll-like receptor (TLR) ligands, and extracellular protein remodeling was amplified in vein grafts versus non-grafted veins. Further, an early (1–3 days) response network in vein grafts was associated with cytoskeletal reorganization and blood coagulation, whereas a late (14–28 days) network associated primarily with chemotaxis. Conversely, a network of proteins involved in metabolism was amplified in normal versus grafted veins. Next, network proximity analysis, integrating data from several human vascular diseases, suggested a link between the mouse vein graft proteome and human arteriovenous fistula disease. Ultimately, experimental data from human vein graft disease would provide greater confidence of target discoveries. Large-scale genetic data, which could provide strong support for causality and directional effects of candidates, are not yet available for human vein graft disease.
Peroxisome proliferator-activated receptor-α (PPARα), a ligand-activated transcription factor known to enhance fatty acid beta-oxidation, and a central protein in the network of proteins enhanced in undisturbed veins, as compared with vein grafts, was selected for further analysis (validation phase, Figure 1). In mouse experiments, silencing macrophage PPARα by nanoparticle delivery of siRNA worsened lesions in vein grafts, whereas treatment with the selective PPARα activator pemafibrate7 slowed onset of vein graft lesion development, reduced lesion accumulation of macrophages, and also protected against detrimental changes in an arteriovenous fistula mouse model.
Next, the authors performed a series of single-cell RNA-sequencing and metabolomics studies in isolated human macrophages stimulated with the TLR4 ligand lipopolysaccharide (LPS) and treated with pemafibrate. These studies suggested that pemafibrate prevents some of the inflammatory effects of LPS in a subpopulation of human macrophages, while increasing genes related to fatty acid oxidation. The metabolomics showed that PPARα activation indeed shifts metabolism from glycolysis to oxidative respiration. Finally, Decano et al. performed a directed regulatory network analysis to investigate the relevance of the in vitro-generated metabolomics data to the vein graft mouse model. These results, based on a small number of mice, appeared to support a direct role of PPARα in vein graft homeostasis.
While the study by Decano and colleagues is an excellent example of use of a non-biased approach for identification of protein targets involved in vein graft disease, further studies will be needed to achieve the goal of using this systems approach for identification of targets in a truly non-biased and human disease-relevant manner. While selection of PPARα for validation studies, rather than one of the more novel proteins identified, did provide important proof-of-concept, PPARα activation by pemafibrate has previously been shown to suppress atherosclerosis and vascular response to injury,8, 9 and to increase fatty acid oxidation and suppress inflammatory activation in macrophages.10 Thus, the selection of PPARα as the target for follow-up studies, as well as the selection of cell type (i.e., macrophages rather smooth muscle cell-derived cell types), the lack of primary discovery in human tissues, and the severe hyperlipidemia of the mouse model introduce biases and limitations to the approach used.
A novel aspect of the study is the demonstration that pemafibrate prevents vein graft disease and arteriovenous fistula pathology in mice. In support, gemfibrozil, an older fibrate, reduced the number of new lesions in venous aortocoronary bypass grafts in a small clinical trial of men with low HDL-cholesterol.11 The large PROMINENT trial (NCT03071692) should report in 2022 and will reveal if pemafibrate will prevent cardiovascular outcomes in subjects with type 2 diabetes and elevated plasma triglycerides.12 Pemafibrate has a higher potency and fewer off-target effects than previously tested fibrates, which have largely failed to prevent CVD.13 Decano et al. selected a dose (0.2 mg/kg body weight/day) of pemafibrate to not lower plasma triglycerides, although this dose did have a triglyceride-lowering effect under fasting conditions. Furthermore, this dose is higher than that used in PROMINENT (0.2 mg twice/day), a dose that lowers triglycerides by approximately 50% in humans.14 It is therefore possible that the beneficial effects of pemafibrate on vein graft lesions in the mice might have been due in part to modulation of circulating lipid species, or to indirect effects of pemafibrate in other tissues, such as increased hepatic expression of the anti-atherogenic FGF21.15
In summary, Decano et al. provide an interesting proof-of-principle supporting the concept that drug targets effective in preventing complex CVD, such as vein graft disease, can be identified by integrative systems approaches. Will pemafibrate be effective in preventing vein graft disease in humans? While we await the results of the PROMINENT trial and before a placebo-controlled clinical trial of pemafibrate is indicated for vein graft disease, much can be done to build on the current work and to delve more deeply into a fully integrated systems approach (Figure 1) to identify novel molecular and cellular targets for vein graft disease in humans.
Sources of funding
Work in the authors’ laboratories is supported by grants from the National Institutes of Health P01HL151328, P01HL092969, R01HL149685, and R35HL150754 (to K.E.B.) and UL1TR001873, K24HL107643, R01HL150359, R01HL132561 and R01HL113147 (to M.P.R.).
Footnotes
Disclosures
None.
References
- 1.Williams JW, Winkels H, Durant CP, Zaitsev K, Ghosheh Y and Ley K. Single Cell RNA Sequencing in Atherosclerosis Research. Circ Res. 2020;126:1112–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB and Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, Ihuegbu CO, Bush EC, Worley J, Vlahos L, Laise P, Solomon RA, Connolly ES, Califano A, Sims PA, Zhang H, Li M and Reilly MP. Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation. 2020;142:2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess DJ. Reaching completion for GTEx. Nat Rev Genet. 2020;21:717. [DOI] [PubMed] [Google Scholar]
- 5.Decano JL, Singh SA, Bueno CG, Lee LH, Halu A, Chelvanambi S, Matamalas JT, Zhang H, Mlynarchik AK, Qiao J, Sharma A, Mukai S, Wang J, Anderson DG, Ozaki CK, Libby P, Aikawa E and Aikawa M. Systems Approach to Discovery of Therapeutic Targets for Vein Graft Disease. PPAR-alpha Pivotally Regulates Metabolism, Activation, and Heterogeneity of Macrophages and Lesion Development. Circulation. 2021;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding H, Douglass EF Jr., Sonabend AM, Mela A, Bose S, Gonzalez C, Canoll PD, Sims PA, Alvarez MJ and Califano A. Quantitative assessment of protein activity in orphan tissues and single cells using the metaVIPER algorithm. Nat Commun. 2018;9:1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fruchart JC, Santos RD, Aguilar-Salinas C, Aikawa M, Al Rasadi K, Amarenco P, Barter PJ, Ceska R, Corsini A, Despres JP, Duriez P, Eckel RH, Ezhov MV, Farnier M, Ginsberg HN, Hermans MP, Ishibashi S, Karpe F, Kodama T, Koenig W, Krempf M, Lim S, Lorenzatti AJ, McPherson R, Nunez-Cortes JM, Nordestgaard BG, Ogawa H, Packard CJ, Plutzky J, Ponte-Negretti CI, Pradhan A, Ray KK, Reiner Z, Ridker PM, Ruscica M, Sadikot S, Shimano H, Sritara P, Stock JK, Su TC, Susekov AV, Tartar A, Taskinen MR, Tenenbaum A, Tokgozoglu LS, Tomlinson B, Tybjaerg-Hansen A, Valensi P, Vrablik M, Wahli W, Watts GF, Yamashita S, Yokote K, Zambon A and Libby P. The selective peroxisome proliferator-activated receptor alpha modulator (SPPARMalpha) paradigm: conceptual framework and therapeutic potential : A consensus statement from the International Atherosclerosis Society (IAS) and the Residual Risk Reduction Initiative (R3i) Foundation. Cardiovasc Diabetol. 2019;18:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hennuyer N, Duplan I, Paquet C, Vanhoutte J, Woitrain E, Touche V, Colin S, Vallez E, Lestavel S, Lefebvre P and Staels B. The novel selective PPARalpha modulator (SPPARMalpha) pemafibrate improves dyslipidemia, enhances reverse cholesterol transport and decreases inflammation and atherosclerosis. Atherosclerosis. 2016;249:200–8. [DOI] [PubMed] [Google Scholar]
- 9.Konishi H, Miyauchi K, Onishi A, Suzuki S, Fuchimoto D, Shitara J, Endo H, Wada H, Doi S, Naito R, Ogita M, Dohi T, Kasai T and Daida H. Effect of pemafibrate (K-877), a novel selective peroxisome proliferator-activated receptor alpha modular (SPPARMalpha), in atherosclerosis model using low density lipoprotein receptor knock-out swine with balloon injury. PLoS One. 2020;15:e0241195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigamonti E, Chinetti-Gbaguidi G and Staels B. Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler Thromb Vasc Biol. 2008;28:1050–9. [DOI] [PubMed] [Google Scholar]
- 11.Frick MH, Syvanne M, Nieminen MS, Kauma H, Majahalme S, Virtanen V, Kesaniemi YA, Pasternack A and Taskinen MR. Prevention of the angiographic progression of coronary and vein-graft atherosclerosis by gemfibrozil after coronary bypass surgery in men with low levels of HDL cholesterol. Lopid Coronary Angiography Trial (LOCAT) Study Group. Circulation. 1997;96:2137–43. [DOI] [PubMed] [Google Scholar]
- 12.Pradhan AD, Paynter NP, Everett BM, Glynn RJ, Amarenco P, Elam M, Ginsberg H, Hiatt WR, Ishibashi S, Koenig W, Nordestgaard BG, Fruchart JC, Libby P and Ridker PM. Rationale and design of the Pemafibrate to Reduce Cardiovascular Outcomes by Reducing Triglycerides in Patients with Diabetes (PROMINENT) study. Am Heart J. 2018;206:80–93. [DOI] [PubMed] [Google Scholar]
- 13.Sandesara PB, Virani SS, Fazio S and Shapiro MD. The Forgotten Lipids: Triglycerides, Remnant Cholesterol, and Atherosclerotic Cardiovascular Disease Risk. Endocr Rev. 2019;40:537–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arai H, Yamashita S, Yokote K, Araki E, Suganami H, Ishibashi S and Group KS. Efficacy and Safety of Pemafibrate Versus Fenofibrate in Patients with High Triglyceride and Low HDL Cholesterol Levels: A Multicenter, Placebo-Controlled, Double-Blind, Randomized Trial. J Atheroscler Thromb. 2018;25:521–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raza-Iqbal S, Tanaka T, Anai M, Inagaki T, Matsumura Y, Ikeda K, Taguchi A, Gonzalez FJ, Sakai J and Kodama T. Transcriptome Analysis of K-877 (a Novel Selective PPARalpha Modulator (SPPARMalpha))-Regulated Genes in Primary Human Hepatocytes and the Mouse Liver. J Atheroscler Thromb. 2015;22:754–72. [DOI] [PMC free article] [PubMed] [Google Scholar]


