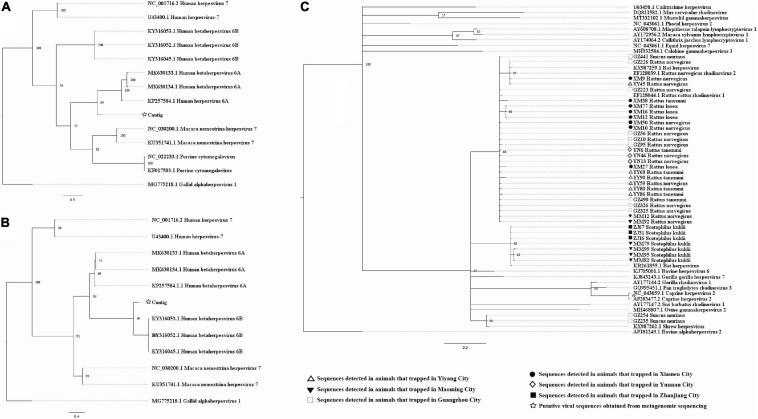
FIGURE 7.
(A) Phylogenetic tree constructed based on potential viral nucleotide sequence (451 bp, IE-2 protein gene) related to human herpesvirus 6A that obtained by using metagenomic sequencing (MrBayes, GTR + G + I nucleotide substitution model). A total of 10 representative sequences belonging to different species within genus Roseolovirus are included for comparison, and one sequences belonging to genus Iltovirus is set as outgroup. Percentages of the posterior probability (PP) values are indicated. (B) Phylogenetic tree constructed based on potential viral nucleotide sequence (245 bp) related to human herpesvirus 6B that obtained by using metagenomic sequencing (MrBayes, GTR + G + I nucleotide substitution model). A total of 12 representative sequences belonging to different species within genus Roseolovirus are included for comparison, and one sequences belonging to genus Iltovirus is set as outgroup. Percentages of the posterior probability (PP) values are indicated. (C) Phylogenetic tree constructed based on PCR-screened nucleotide sequences (144 bp) of herpesviruses from urban rats, bats, and house shrews (MrBayes, GTR + G + I nucleotide substitution model). A total of 23 representative sequences (DPOL gene region) are included for comparison, including 20 sequences belonging to different genera within subfamily Gammaherpesvirinae, one bat herpesvirus sequence, one shrew herpesvirus sequence, and one alphaherpesvirus sequence that is set as outgroup. Percentages of the posterior probability (PP) values are indicated.