Figure 5.
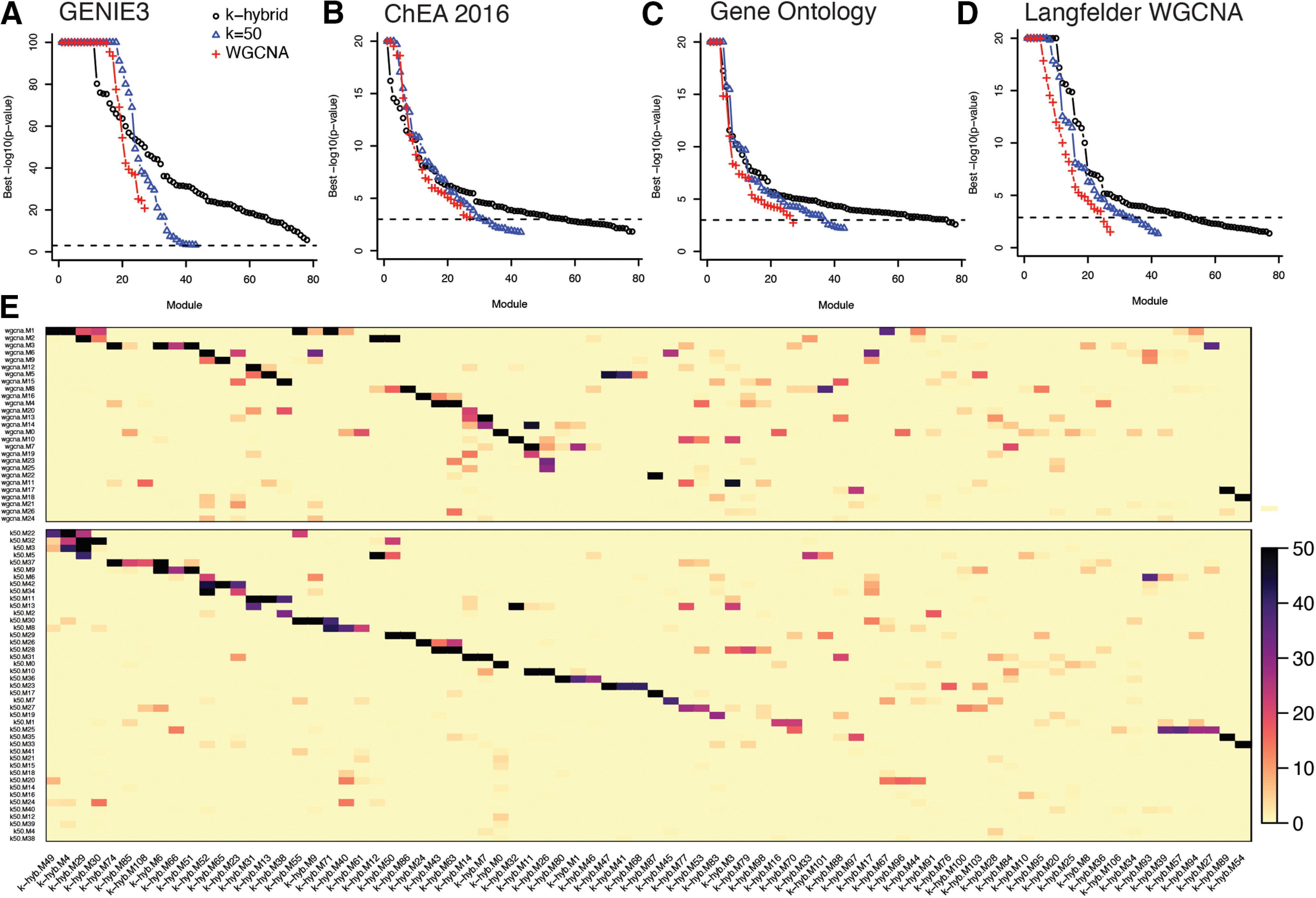
Validation and comparison of gene co-expression modules. A, Enrichment of gene co-expression modules for TF-target regulons derived with GENIE3 (Huynh-Thu et al., 2010). B, Enrichment of gene co-expression modules for TF-target regulons from ChIP-seq experiments in the ChEA 2016 database (Lachmann et al., 2010). C, Enrichment of gene co-expression modules for Gene Ontology terms. D, Enrichment of gene co-expression modules for published gene co-expression modules derived bulk RNA seq of HTT knock-in mice (Langfelder et al., 2016). The points in A–D indicate the -log10(p value) for the most significant regulon or GO term for each module, with the modules ranked from strongest to weakest enrichment. E, Overlap between gene co-expression modules derived with WGCNA, k-means (k = 50), and the k-means hybrid approach.
