Fig. 2. PRV preferentially interacts with open and active chromatin regions in the swine genome.
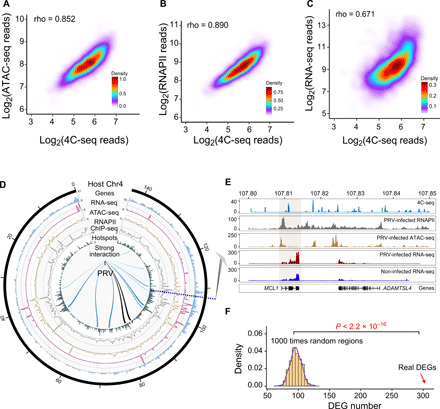
(A) Density plot demonstrating the positive correlation between ATAC-seq reads and PRV-host 4C reads in PRV-infected cells. (B) Density plot showing the positive correlation between RNAPII ChIP-seq reads and PRV-host 4C reads. (C) Density plot showing the positive correlation between RNA sequencing (RNA-seq) reads and PRV-host 4C reads. (D) Circos plot of the integrated 4C-seq data, ATAC-seq, RNAPII ChIP-seq, and RNA-seq data on chromosome 4. (E) Integrated sequencing tracks for the 4C-seq data, ATAC-seq, RNAPII ChIP-seq, and RNA-seq data of the MCL1 gene. (F) Number of differentially expressed genes (DEGs) related to the random regions and interaction regions, respectively, indicating that DEGs are enriched in the interaction hotspots. P < 2.2 × 10−16 from the Student’s t test.
