Fig. 3. DNA binding protein RUNX1 mediates the PRV-host chromatin interaction.
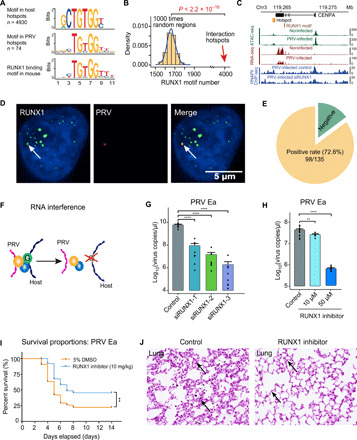
(A) Significant protein binding motifs in both PRV and host cell interaction hotspots and the potential DNA binding protein RUNX1. (B) Number of RUNX1 motifs in random regions and interaction hotspots, respectively. (C) Integrated sequencing tracks for the RUNX1 motif, 4C-seq data, ATAC-seq, RNA-seq data, and RNAPII ChIP-seq in CENPA gene locus. (D) Colocalization of the RUNX1 protein immunofluorescence signal and PRV genome DNA FISH (HCR) signal. (E) PRV genome and RUNX1 protein colocalized in 72.6% (98 of 135) of the tested cells. (F) Schematic illustration of the RUNX1 RNA interference effect on PRV-host chromatin interaction. (G) Significant decrease of PRV copy number after RUNX1 interference in PRV-infected PK15 cells. (H) PRV copy number in PRV-infected PK15 cells treated with different concentrations of RUNX1 inhibitor (Ro 5-3335). (I) Survival curves of the PRV-infected C57BL/6 mice treated with Ro 5-3335 and 5% dimethyl sulfoxide (DMSO; control); n = 42 per group. (J) Histological staining of the lung with or without the RUNX1 inhibitor Ro 5-3335 treatment after PRV Ea infection, respectively. Arrows show the thickened alveolar septum in control group. **P < 0.01 and ****P < 0.0001 from Student’s t test. Error bars represent SD from eight independent experiments.
