Fig. 4. PRV hijacks host RNAPII via RUNX1 for its transcription and replication.
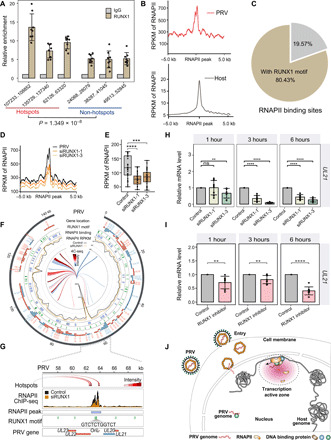
(A) RUNX1 ChIP-qPCR assay demonstrated the enrichment of RUNX1 in the hotspot and non-hotspot regions in the PRV genome [normalized with immunoglobulin G (IgG) control and input]. (B) Average read coverage of RNAPII peaks in the PRV and host genomes. RPKM, reads per kilobase per million mapped reads. (C) 80.43% of the RNAPII peaks were located around the RUNX1 binding motif in PRV genome (<2 kb). (D) Average read coverage of RNAPII peaks in the PRV genome with/without RUNX1 RNA interference. (E) Box plot of RNAPII occupancy on PRV genome with/without RUNX1 RNA interference. (F) Distribution of RNAPII binding sites, RNAPII occupancy, RUNX1 motif, and genes in the PRV genome with/without RUNX1 RNA interference. (G) Distribution of RNAPII binding sites, RNAPII occupancy, RUNX1 motifs, and genes in OriL region (around UL21 locus) of PRV genome with/without RUNX1 RNA interference. (H) Expression levels of UL21 at 1, 3, and 6 hours with/without RUNX1 RNA interference. (I) Expression levels of UL21 at 1, 3, and 6 hours following treatment with RUNX1 inhibitor Ro 5-3335. (J) Model of chromatin interaction between pseudorabies viral genome and host cell genome. ns, not significant (P > 0.05); **P < 0.01, ***P < 0.001, and ****P < 0.0001 from the Student’s t test. Error bars represent SD from eight independent experiments.
