Abstract
We aimed to validate oxygen extraction fraction (OEF) estimations by quantitative susceptibility mapping plus quantitative blood oxygen-level dependence (QSM+qBOLD, or QQ) using 15O-PET. In ten healthy adult brains, PET and MRI were acquired simultaneously on a PET/MR scanner. PET was acquired using C[15O], O[15O], and H2[15O]. Image-derived arterial input functions and standard models of oxygen metabolism provided quantification of PET. MRI included T1-weighted imaging, time-of-flight angiography, and multi-echo gradient-echo imaging that was processed for QQ. Region of interest (ROI) analyses compared PET OEF and QQ OEF. In ROI analyses, the averaged OEF differences between PET and QQ were generally small and statistically insignificant. For whole brains, the average and standard deviation of OEF was 32.8 ± 6.7% for PET; OEF was 34.2 ± 2.6% for QQ. Bland-Altman plots quantified agreement between PET OEF and QQ OEF. The interval between the 95% limits of agreement was 16.9 ± 4.0% for whole brains. Our validation study suggests that respiratory challenge-free QQ-OEF mapping may be useful for non-invasive clinical assessment of regional OEF impairment.
Keywords: Oxygen extraction fraction, positron emission tomography, quantitative susceptibility mapping, quantitative blood oxygenation level-dependent imaging, QSM+qBOLD
Introduction
Regional oxygen extraction fraction (OEF) is an essential biomarker for investigating tissue vulnerability and function in various diseases such as stroke,1–4 cerebral tumors,5 and Alzheimer’s Disease.6 Positron emission tomography (PET) with 15O tracers is the reference standard for quantitative mapping of OEF.7–9 Using tracer kinetic modeling of 15O tracers, PET has been used to map OEF in healthy subjects,8,10 various diseases including stroke2,11–15 and Huntington’s disease.16 An image-derived arterial input function method was further introduced for 15O PET imaging using PET/MR.17,18 However, PET with 15O has not been widely used in clinical settings because 15O tracers with 122-second half-lives must be produced by a cyclotron within the PET facility.8 This has substantially limited 15O PET availability.
In contrast, with widely available MR scanners, tissue cerebral oxygen consumption can be estimated by modeling conversion of diamagnetic oxyheme into paramagnetic deoxyheme in the vasculature. OEF can be estimated from MRI signal magnitudes by methods such as T2-Relaxation-Under-Spin-Tagging (TRUST),19–21 quantitative BOLD (qBOLD),22–26 quantitative imaging of extraction of oxygen and tissue consumption (QUIXOTIC),27 and calibrated BOLD.28–31 OEF can also be estimated from MRI signal phase by methods such as whole-brain susceptometry-based oximetry,32,33 and quantitative susceptibility mapping (QSM)34 methods using macrovascular15,35,36 or microvascular models.37–39
A promising, recently reported OEF mapping method uses a comprehensive MR signal model incorporating both signal phase and magnitude (QSM+qBOLD, or QQ).40 QQ estimates OEF maps from multi-echo gradient (mGRE) data alone. It does so without burdensome gas inhalation or respiratory-control procedures. The robustness of QQ OEF has been significantly improved by introduction of an unsupervised machine learning method, cluster analysis of time evolution, which may enable clinically practical use of the QQ OEF mapping method.41 Accordingly, the purpose of this study is to validate QQ OEF measurements as compared to reference standard 15O PET OEF measurements in healthy adults.
Materials and methods
Data acquisition
Ten healthy subjects (8 females, age 43 ± 20 years) underwent MRI and PET. The study was approved by Washington University Human Research Protection Office and Institutional Review Board, and written informed consent was obtained from all individuals in accordance with the ethical standards of the Helsinki Declaration of 1975 and its later amendments.
All subjects were imaged on a PET/MR system (Siemens Biograph 3 T mMR, Erlangen, Germany). Anatomical MRI images were first acquired. PET data was acquired with sequential administrations of C[15O], O[15O], H2[15O], C[15O], O[15O], and H2[15O]. There was greater than six half-lives between consecutive administrations of 15O tracers. Prescribed doses were 15-37 mCi for C[15O] admixed in room air, 15-37 mCi of O[15O] in room air and 25 mCi of H2[15O] injected intravenously as a bolus. During PET, MRI was acquired simultaneously. MRI included structural Magnetization Prepared Rapid Gradient Echo (MPRAGE) and time-of-flight MR angiography (TOF-MRA) that were used for anatomic registration and for calculating an image-derived arterial input function (IDAIF), and duplicate sets of mGRE sequences for QQ that were aimed to temporally coincide with O[15O] scans. (Due to technical issue, in MRI, only a single mGRE was acquired on subject 9 and, in PET, one C[15O] scan was missed on subject 1 and 6, and one H2[15O] scan was missed on subject 7.)
Acquisitions of PET list mode data began at least 60 seconds prior to administration of each tracer and the PET scan duration included 720 seconds of listmode packets. Attenuation mu-maps were synthesized from MPRAGE using the method of Burgos, et al.42 Dynamic imaging frames were reconstructed with four iterations of ordered sets of expectation maximization and isotropic Gaussian filtering at 4.3 mm full-width half-maximum (FWHM) using the NiftyPET software platform.43
MRI imaging parameters were as follows. T1-MPRAGE was acquired for T1-weighted anatomical image (T1w): TR = 2400ms, TE = 2.97 ms, TI =1000ms, flip angle = 8°, and voxel size = 0.95 × 0.95 ×1 mm3. TOF-MRA was acquired for IDAIF: TR = 22ms, TE = 3.94 ms, flip angle = 18°, and voxel size = 0.57 × 0.57 × 0.70 mm3. mGRE was acquired for QQ-OEF: TR= 33 ms, TE1/ΔTE/TE10 = 4.7/2.5/28.4 ms, flip angle 15°, bandwidth = 465 Hz/pixel, voxel size =0.94 × 0.94 × 3 mm3.
Data processing
QQ-OEF mapping from mGRE data: The QQ model estimates oxygen extraction fraction based on the venous deoxyheme-dependent signal in mGRE signal phase using QSM and signal magnitude using qBOLD.41 The QSM modeling considers that voxel-wise susceptibility is the sum of three components: non-blood tissue susceptibility (χnb), the plasma susceptibility, and the hemoglobin susceptibility.37,39,44 The hemoglobin susceptibility is mainly determined by venous blood volume (v) and venous oxygenation (Y). For instance, the hemoglobin susceptibility increases as v increases and Y decreases. The qBOLD modeling distinguishes the mGRE magnitude signal decay into three contributions: irreversible microscopic field contribution by spin-spin interaction (R2 effect, nm scale), mesoscopic field contribution by the susceptibility difference between blood and surrounding tissue (FBOLD effect, µm scale), and macroscopic field contribution from air-tissue interfaces (mm scale).23,45 The FBOLD effect is induced by v, Y, and χnb. For instance, the FBOLD effect increases as v increases and Y and χnb decrease. As the QSM and qBOLD commonly have v, Y, and χnb, OEF (=1−Y/Ya where Ya: arteriole oxygenation) can be estimated by the combined model of QSM+qBOLD (QQ).
The two inputs for QQ are voxel-wise susceptibility and mGRE magnitude signal. First, the susceptibility was estimated as follows. The total field was obtained with a linear fit of the mGRE phase.46 The local field was subsequently estimated by the projection onto dipole field (PDF) method.47 Susceptibility was then computed by the Morphology Enabled Dipole Inversion with automatic uniform cerebrospinal fluid zero reference (MEDI + 0) algorithm.48–51 Based on the obtained susceptibility and mGRE magnitude, OEF was estimated using QQ.40,41
Since the inversion of QQ is involved with nonconvex optimization due to the coupling of v and Y, it is very sensitive to the measurement noise. For robust OEF reconstruction against noise, cluster analysis of time evolution (CAT) was used.41 The basic idea of CAT is that the voxels with similar mGRE signal decay can be grouped into a cluster and be assumed to have similar model parameter values including OEF. Consequently, signal-to-noise ratio is expected to be increased significantly by averaging over numerous voxels in a cluster. The QQ-OEF was registered to the T1w images using the FSL FLIRT algorithm.52,53
15O PET-OEF mapping: PET-OEF was estimated using two compartmental tracer kinetic modeling8 and IDAIF based on the PET/MR hybrid scanner approach.17 First, cross modality registration was performed, e.g. PET to T1w and TOF-MRA to T1w.54,55 PET images were smoothed to a common resolution of 8 mm FWHM to minimize inter-scan differences in PET resolution.56 Time-activity curves (TACs) were obtained in regional ROIs identified by FreeSurfer57 based upon the MPRAGE images and in arterial ROIs determined based on TOF-MRA using an adaptive segmentation algorithm,18,58,59 respectively.
Using the tracer kinetic modeling with these TACs, CBF and CBV were estimated from the 15O-water scans9,60 and 15O-carbon monoxide scans,61 respectively. OEF was finally estimated from the 15O-oxygen scans in conjunction with calculated CBF and CBV images.58 For robust PET-OEF estimation, nonlinear curve fitting was only performed for the estimation of IDAIFs for the 15O-water scans.18 For 15O-Carbon monoxide and 15O-oxygen estimation, the IDAIF was estimated based on the measured arterial and background ROI TAC,17 where no curve fitting was involved. For CBV estimation, although the 15O-Carbon monoxide was acquired as multi-frame dynamic emission scans, the steady-state portion of the scan (after 2 minutes post initiation of inhalation) was numerically integrated to generate a single frame static scan, and the voxel-wise signal was estimated as the ratio of voxel intensity to the arterial signal intensity. To avoid performing model fitting with extremely noisy TACs in the estimation of voxel-wise CBF maps and subsequent OEF maps, the linearized version of the original CBF and OEF model was used.17
ROI analysis
QQ-OEF was smoothed with 3 D Gaussian filtering to match that applied to PET-OEF (FWHM = 8 mm). To compare QQ and PET, QQ- and PET-OEF maps were averaged over the two scans (scan-rescan). Comparisons of OEF measures between QQ-OEF and PET-OEF were performed in the whole brain and regional ROIs: cortical gray matter (CGM), frontal, temporal, parietal, and occipital lobe of CGM, white matter (WM), and deep gray matter (DGM) regions (Thalamus, Caudate, Putamen, and Pallidum). The OEF values were presented as mean ± standard deviation. Paired t-tests estimated significant differences between QQ-OEF and PET-OEF. A P value less than 0.01 was considered significant. The Lilliefors test confirmed the normality of each ROI data distribution (all p-values > 0.025). Bland-Altman (BA) plots were constructed based on the OEF average in regional ROIs. For a non-uniform difference in BA plots, a regression approach was used to obtain the 95% limits of agreement with considering a linear trend as in Figures 2(a), (c) and 4(a), (c).62 Each ROI mask was identified by FreeSurfer57 analysis of the T1w MPRAGE sequence and overlapped with a QQ reconstruction whole brain mask registered to the T1w MPRAGE beforehand (Supporting Information Figure S1). The voxels with extremely high OEF value (>90%) observed on PET-OEF were excluded from ROI analysis. Those were not physiologic and likely to be caused by artefacts, such as misalignments between component scans that lead to errors near vasculature, and bone/CSF or attenuation/scatter correction errors that lead to hot spots.
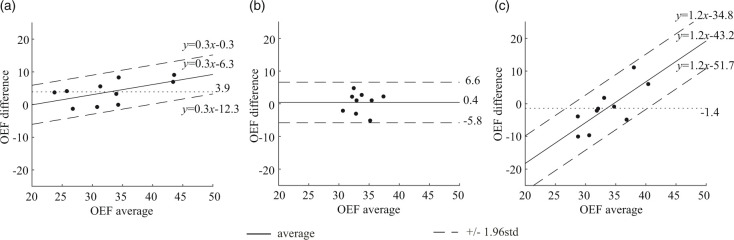
Figure 2.
Bland-Altman plots comparing OEF values in whole brain between PET and QQ scans. (a) PET scan 1 vs. PET scan 2. (b) QQ scan 1 vs. QQ scan 2. (C) PET average vs. QQ average. PET and QQ show small scan to rescan variations (average OEF difference: 3.9%, p < 0.009 for PET and 0.4%, p = 0.7 for QQ). The average difference between PET and QQ is not significant (−1.4%, p = 0.5). The OEF agreement interval (distance between the two dashed line) between PET and QQ average is similar to the one between scan-rescan within each method. The unit in the x- and y-axis is %.
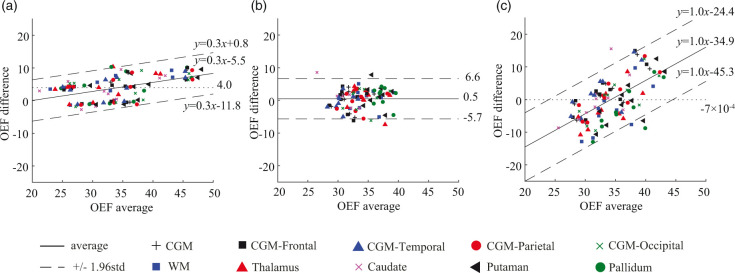
Figure 4.
Bland-Altman plots comparing OEF values in regional ROIs between PET and QQ scans. (a) PET scan 1 vs. PET scan 2. (b) QQ scan 1 vs. QQ scan 2. (c) PET average vs. QQ average. A small and statistically not significant bias in mean regional OEF difference between PET and QQ averages was found (dotted line in (c)). The OEF agreement interval (mean ± 1.96std) between two methods was comparable to the one between scan-rescan within each method. The unit in the x- and y-axis is %.
Results
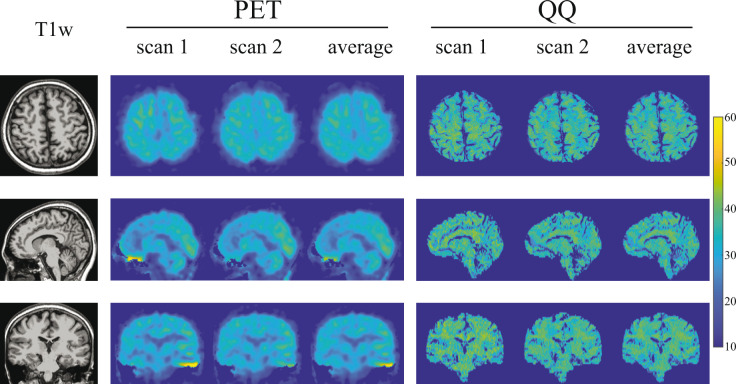
Figure 1 shows, for an exemplar subject, representative OEF maps generated by PET and QQ. Excluding boundary voxels where division by small estimators of tracer dynamics create unstable PET point-estimates, both PET and QQ provide OEF estimates of 30.8 ± 4.0% for PET and 32.9 ± 4.4% for QQ. OEF maps in all the 10 subjects are shown in Supporting Information Figures S2, S3, and S4.
Figure 1.
OEF maps from PET and QQ in axial, sagittal, and coronal views in a subject. Both PET and QQ show uniform OEF maps and good agreement between scans and methods.
Figure 2 shows Bland-Altman residual plots of whole-brain OEF. PET had scan-rescan variations (Figure 2(a)). Averaging over all subjects, the OEF difference between scans was 3.9% (p = 0.009), marked by a dotted line. In QQ, averaging over all subjects, the OEF difference between scans was 0.4% (p = 0.7), similarly marked by a dotted line (Figure 2(b)). QQ showed similar average whole brain OEF values when compared to PET (Figure 2(c) and Table 1): 32.8 ± 6.7% on PET and 34.2 ± 2.6% on QQ (p = 0.5). When analyzing the scan-rescan results separately, the interval of the 95% limits of agreement between PET and QQ was similar to that between scan-rescan for each method: 16.9 ± 4.0% for PET and QQ average, 12.0 ± 2.8% for PET scan 1 and 2, and 12.4 ± 3.1% for QQ scan 1 and 2. Figure 2(a) and (c) shows a linear trend (p-values < 0.0001).
Table 1.
Average and standard deviation of whole brain OEF.
|
PET |
|
|||||
|---|---|---|---|---|---|---|
| Subject | Scan 1 | Scan 2 | average | Scan 1 | Scan 2 | Average |
| 1 | 46.9 ± 5.4 | 39.9 ± 5.0 | 43.4 ± 4.9 | 38.5 ± 5.3 | 36.3 ± 5.2 | 37.4 ± 5.1 |
| 2 | 35.6 ± 5.9 | 32.3 ± 4.4 | 34.0 ± 4.8 | 33.3 ± 3.5 | 31.1 ± 3.3 | 32.2 ± 3.1 |
| 3 | 25.6 ± 3.8 | 21.9 ± 3.2 | 23.7 ± 3.3 | 35.1 ± 5.6 | 32.4 ± 4.8 | 33.8 ± 5.0 |
| 4 | 26.1 ± 3.6 | 27.4 ± 3.2 | 26.8 ± 3.2 | 29.6 ± 3.1 | 31.7 ± 3.9 | 30.7 ± 3.4 |
| 5 | 27.8 ± 3.2 | 23.7 ± 3.3 | 25.8 ± 3.1 | 35.9 ± 4.1 | 34.9 ± 4.5 | 35.4 ± 4.2 |
| 6 | 34.1 ± 4.1 | 28.5 ± 4.4 | 31.3 ± 3.7 | 31.3 ± 4.0 | 34.4 ± 1.1 | 32.8 ± 2.4 |
| 7 | 30.4 ± 4.3 | 31.2 ± 3.9 | 30.8 ± 4.0 | 33.4 ± 4.5 | 32.4 ± 5.0 | 32.9 ± 4.4 |
| 8 | 34.3 ± 3.8 | 34.3 ± 4.1 | 34.3 ± 3.8 | 32.6 ± 4.1 | 37.8 ± 5.9 | 35.2 ± 4.6 |
| 9 | 38.5 ± 4.5 | 30.2 ± 3.5 | 34.4 ± 3.6 | 39.2 ± 5.4 | N.A. | 39.2 ± 5.4 |
| 10 | 48.1 ± 4.8 | 39.0 ± 4.0 | 43.6 ± 4.0 | 34.8 ± 3.8 | 30.1 ± 5.4 | 32.5 ± 4.0 |
| avg ± std | 34.3 ± 8.3 | 30.9 ± 6.2 | 32.8 ± 6.7 | 33.8 ± 2.6 | 33.4 ± 2.5 | 34.2 ± 2.6 |
Note: The unit of OEF is percent. No significant difference was found between PET and QQ average (p = 0.5). PET and QQ show small scan to rescan variations (p < 0.009 for PET and p = 0.7 for QQ). N.A. indicates that the data is not available.
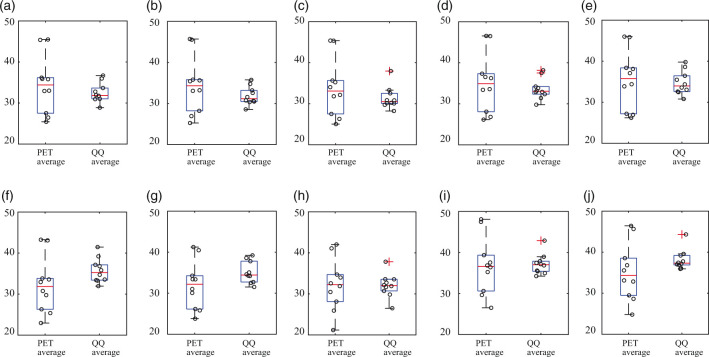
Figure 3 compares PET and QQ estimates of OEF regionally averaged by cortical gray matter (CGM), white matter (WM), and deep gray matter (DGM). PET and QQ provided similar regional OEF values: The averaged OEF values, respectively for PET and QQ, were 34.4 ± 7.0% and 32.5 ± 2.4% in CGM, 34.5 ± 7.0% and 31.9 ± 2.2% in CGM-Frontal, 33.8 ± 7.1% and 31.4 ± 2.7% in CGM-Temporal, 35.1 ± 7.3% and 33.6 ± 2.6% in CGM-Parietal, 35.4 ± 7.2% and 34.7 ± 2.9% in CGM-Occipital, 32.2 ± 6.8 and 35.7 ± 3.0% in WM, 32.0 ± 5.9 and 35.1 ± 2.7% in Thalamus, 32.2 ± 6.4 and 32.1 ± 2.9% in Caudate, 36.8 ± 7.0 and 37.1 ± 2.5% in Putamen, and 35.2 ± 7.0 and 38.1 ± 2.5% in Pallidum. The difference between PET and QQ average was not significant in any of these ROIs (all p-values > 0.12). In CGMs, both PET and QQ showed significantly higher OEF values in CGM-Occipital than CGM average (p = 0.004 for PET and p < 0.0001 for QQ).
Figure 3.
OEF comparison in cortical gray matters (a–e), white matter (f), and deep gray matters (g–j) among PET and QQ average. No significant difference was found between PET and QQ (p > 0.12, paired t-test). The unit in y-axis is %. Red line, blue box, black whisker, and red cross, black circle indicates median value, interquartile range, the range extending to 1.5 of the interquartile range, outlier beyond the whisker range, and individual subject value.
Figure 4 shows the Bland-Altman plots of the regionally averaged OEF values in CGM, WM, and DGM. PET and QQ had a small scan-rescan variation: average OEF difference = 4.0% (p = 0.009) for PET (Figure 4(a)) and 0.5% (p = 0.6) for QQ (Figure 4(b)). The mean regional OEF difference was not significant between PET and QQ averages, −7 × 10−4% (p = 0.97) (dotted line in Figure 4(c)). The interval of the 95% limits of agreement between the two methods across tissue types was comparable to the one between scan-rescan within each method: 20.9 ± 1.5% for PET and QQ average, 12.7 ± 0.9% for PET scan 1 and 2, and 12.3 ± 0.9% for QQ scan 1 and 2. Figure 4(a) and (c) shows a linear trend (p-values < 0.0001).
Discussion
Our data indicate that gradient echo MRI-based QSM+qBOLD (QQ)-OEF mapping is valid against reference 15O PET-OEF mapping in healthy adults; the QQ method provides similar OEF values both globally and regionally when compared to 15O PET. Further validation of challenge-free QQ-OEF mapping in clinical settings is now warranted, as a non-invasive and more accessible assessment of regional OEF impairment than the reference standard 15O PET-OEF technique.
QQ-OEF mapping may be particularly valuable for more widespread and repeated evaluation of cerebral oxygen deficiency causing brain tissue vulnerability or injury in various brain disorders, such as ischemic stroke,63 Alzheimer’s disease (AD),64,65 and multiple sclerosis.66 For instance, in stroke therapy, it is critical to identify salvageable ischemic tissue to determine treatment such as mechanical thrombectomy. Inadequacies of methods in current clinical use, e.g. problematic ischemic core definition by CBF reduction67 and variability in diffusion perfusion mismatch,68–70 can be overcome by an accurate regional OEF mapping method. Another example is measurement of altered brain aerobic glycolysis in AD.71 This measurement partly depends on quantitative measurement of oxygen metabolism, which is particularly difficult to perform with PET in the very large cohorts of individuals comprising current AD studies.
In this study, both PET and QQ showed fairly uniform OEF maps over the brain (Figure 1), which is in line with previous PET and MRI studies.8,10,23,27,72 Global and CGM OEF values estimated with the two methods were not significantly different and agree with prior OEF values obtained from PET, e.g. 35 ± 7% to 40 ± 9%,10,16,72,73 from calibrated BOLD, e.g. 35 ± 4% to 44 ± 14%,30,74–80 and from QSM, e.g. 29 ± 3% to 50 ± 5%.15,36–41,74 Also, the slightly higher OEF in the occipital lobe than CGM average agrees with prior PET literature.72
Compared to PET-OEF, QQ-OEF showed smaller inter-subject variation–average coefficient of variation (COV) in whole brain = 7.6% for QQ vs 20.5% for PET–though their group averages were close (Table 1). This PET inter-subject COV is consistent with similar inter-subject variability for OEF, CBF, and CMRO2 reported in previous PET studies.10,16,17 Though a physiologic reason for this variability is possible, the variability might also arise from various complexities in PET data acquisition and processing. For instance, as PET-OEF estimation depends on CBF and CBV calculated from two independent PET scans with different tracers, CBF and CBV variability may contribute to the OEF variability.8,11 PET-OEF estimation relies on subject inhalation of O[15O] and C[15O] gases, which might introduce another source of inter-subject variability. Also, the process of arterial input function (AIF) estimation needed for quantitative PET analysis is complex, the IDAIF used in this study is sensitive to PET-MRI registration uncertainty, and the resultant variability in AIF estimation may contribute to additional variability in OEF estimation.17 In addition, MR-based attenuation and scatter correction81 used in this study can lead to spatially varying and individual-dependent PET signal variation,82 further contributing to OEF estimation variability.
Regional ROI analysis showed no significant OEF difference between PET and QQ scans in CGM, WM, and DGM (Figure 3). Average OEF maps from PET and QQ were both similarly largely uniform (Figure 1) and had similar global OEF values between the two methods (Figure 2 and Table 1). The within-subject standard deviation of the whole-brain OEF, e.g. 4.9% in the PET average of Subject 1 in Table 1, might be caused by real physiological variation and/or various complexities in data acquisition and processing, e.g. the sensitivity of the IDAIF to PET-MRI registration uncertainty in PET-OEF processing.17 Interestingly, both methods independently demonstrated a slightly but significantly higher OEF in the occipital lobes, confirming that this is likely a true physiologic finding.
To investigate if the usage of CAT affects the concordance of QQ- and PET-OEF, QQ-OEF without CAT was processed by following the original QQ paper40 with v initialized to that used in the QQ-CAT paper41 and showed significant difference from PET-OEF, e.g. 38.5 ± 2.1% vs. 32.8 ± 6.7% (p = 0.02, paired t-test) for the global OEF. This suggests that the usage of CAT seems critical for the QQ- and PET-OEF concordance, which may be related to that the measurement error propagation into the OEF was reduced significantly by using CAT in the QQ model.41
The intra-subject variability existed from scan 1 to scan 2 in PET-OEF. Six out of 10 subjects (Subject 1, 3, 5, 6, 9, and 10) showed a larger than 10% global OEF decrease in Scan 2 compared to Scan 1, whereas 2 subjects (Subject 4 and 7) showed a slight increase, 2∼5% (Table 1). We believe that the deviation between two scans are potentially both artefact-related and physiologic.
Regarding the possibility of artefact, although PET-OEF was estimated twice for all 10 participants, three of them had only one 15O-water or 15O-Carbon monoxide scan, and a single CBF/CBV map was used to estimate the OEF for both 15O-Oxygen scans. Biological fluctuations in CBF/CBV could lead to the biases in OEF. Also, misalignment in the component scans could lead to inaccurate OEF estimations. In addition, the PET in our PET/MR scanner was inferior to traditional PET scanners for 15O PET imaging with suboptimal attenuation and scatter correction, lack of 2 D acquisition mode, difficulty in physically limiting motion artifact, difficulties in getting the inhalation tube to the participants mouth, and unpleasant environment relative to traditional PET scanners due to the smaller and deeper bore size.
Regarding the possibility of real physiologic change, the OEF difference between the two scans in PET correlates with that in QQ, e.g. (R-square = 0.35, p = 0.046) for the whole brain (Table 1, excluding Subject 9 without Scan 2 for QQ). The same trend of change between the two scans on both PET-OEF and QQ-OEF simultaneously acquired on PET/MR might be caused by real physiologic change.
QQ-OEF showed a trend of being greater than PET-OEF (not statistically significant) in white matter and some deep gray nuclei such as Thalamus and Pallidum. It is possible that QQ-OEF is overestimated in those regions. The qBOLD in QQ model assumes two source compartments within a voxel: iron in venous structure (dexoyhemoglobin) and diffusively distributed source (medium), e.g. non-blood tissue protein or ferratin. If the other structured (non-diffusive) strong sources exist, such as myelin in white matter and structured ferratin in deep gray matter, they would induce additional intravoxel field variation, which may lead to greater MRI signal decay than the sole deoxyhemoglobin effects of venous blood. Consequently, QQ-OEF might be overestimated in white matter and some deep gray nuclei.
For the uncertainty analysis on QQ-OEF, we performed a numerical simulation to investigate how much measurement noise propagates into OEF estimation, similarly to the Numerical simulation 2 in the QQ-CAT paper.41 First, mGRE magnitude signal and the susceptibility values were simulated based on the QQ model. The input was the QQ-CAT result of a subject in this study (ground truth). Gaussian noise was then added to the mGRE signal and the susceptibility values to obtain a realistic SNR of 50. The simulated data was processed by the QQ with CAT. This process was repeated for 10 times with additional random gaussian noise at the same noise level (SNR 50). For accuracy measurement, the mean absolute error (MAE) between the average OEF map over the 10 trials and the OEF ground truth was calculated: where i: the voxel index, j: the trial index, : the number of voxels : the number of trials (=10). For precision measurement, the mean standard deviation (MSD) of OEFs among the 10 trials was obtained: . QQ-CAT provides a great accuracy (and high precision ( %) in the OEF estimation. For PET-OEF estimation, the error estimation was not performed as the linearized version of the original OEF model17 was used to avoid nonlinear curve fitting for robust OEF estimation.
We checked how well QQ- and PET-OEF estimates are supported by data (goodness of fit). For QQ-OEF estimation, the relative residual of QSM and qBOLD term in QQ model were calculated: where is the voxel-wise susceptibility and / where S is the mGRE magnitude signal. and (n = 19, two measurements per subject except for one measurement in subject 9). This indicates that the residual norm is substantially lower than the data norm (<5% for QSM and <11% for qBOLD). In PET-OEF estimation, curve fitting was only performed for IDAIF estimation in the 15O-water scans. In an example of total cortical gray matter (one of the ROIs used in the IDAIF modeling), the coefficient of determination was 0.999.
A limitation of this study is that it is not clear which level of sensitivity is required for this technique to be clinically useful. However, the preliminary chronic ischemic stroke patient cases in the QQ-CAT paper showed that QQ-CAT was sensitive enough to capture low OEF regions, which agreed well with DWI-defined ischemic lesions.41 In addition, the healthy subjects in this study showed generally uniform OEF maps without any extreme values, which indicates that no significant false positive error is induced from the QQ-CAT.
To investigate the sensitivity of QQ to OEF abnormalities quantitatively, we performed the same simulation as used in the uncertainty analysis above, with one difference that the ground truth was a simulated acute ipsilateral ischemic stroke patient brain where the OEF map was generally uniform and had regionally higher OEF ischemic lesions. The ground truth ratio of the average OEF in the lesion to the contralateral side () was 1.26 (43.2%/34.2%). QQ-OEF could capture the ratio, 1.27 (43.8%/34.4%) with negligible bias (MAE= %) and great precision (MSD = 4.9 ×10−17%). This result indicates that QQ-CAT can capture such high OEF lesions at SNR 50 reliably, which suggests that the QQ-CAT may be adequate for clinical application, such as lesion investigation in acute ischemic stroke patients where the ratio of OEF in the lesion to contralateral side has been reported to be ∼1.2.83 However, the QQ-CAT should be validated with the reference PET in clinical patients where regional OEF abnormalities are expected.
In conclusion, our study suggests that in healthy adults the QQ model generates whole brain and regional OEF estimates in agreement with the current gold standard OEF estimation by 15O PET. The noninvasive challenge-free gradient echo MRI based QQ OEF mapping is now poised for further evaluation in patients where OEF is likely to be regionally affected.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X20973951 for Cerebral oxygen extraction fraction (OEF): Comparison of challenge-free gradient echo QSM+qBOLD (QQ) with 15O PET in healthy adults by Junghun Cho, John Lee, Hongyu An, Manu S Goyal, Yi Su and Yi Wang in Journal of Cerebral Blood Flow & Metabolism
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported in part by NIH R21EB024366, R01AG057536, R01NS090464, R01NS095562, R01NS105144, S10OD021782, R01AG057536, 1R01NS082561, 1P30NS098577.
Declaration of conflicting interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: JC and YW are in the inventor list on OEF related patent application of Cornell University, and YW owns equity of Medimagemetric LLC. The authors declared no other potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: All authors (JC, JL, HA, MG, YS, and YW) made substantial contribution to the concept and design, data acquisition, or data analysis and interpretation. All authors assisted in drafting or revising the article. Final approval of the submitted manuscript was provided by all authors.
Supplementary material: Supplementary material for this article is available online.
ORCID iDs: Junghun Cho https://orcid.org/0000-0002-0826-5463
Manu S Goyal https://orcid.org/0000-0003-1970-4270
References
- 1.Derdeyn CP, Yundt KD, Videen TO, et al. Increased oxygen extraction fraction is associated with prior ischemic events in patients with carotid occlusion. Stroke 1998; 29: 754–758. [DOI] [PubMed] [Google Scholar]
- 2.Yamauchi H, Fukuyama H, Nagahama Y, et al. Significance of increased oxygen extraction fraction in five-year prognosis of major cerebral arterial occlusive diseases. J Nucl Med 1999; 40: 1992–1998. [PubMed] [Google Scholar]
- 3.Derdeyn CP, Videen TO, Yundt KD, et al. Variability of cerebral blood volume and oxygen extraction: stages of cerebral haemodynamic impairment revisited. Brain 2002; 125: 595–607. [DOI] [PubMed] [Google Scholar]
- 4.Gupta A, Chazen JL, Hartman M, et al. Cerebrovascular reserve and stroke risk in patients with carotid stenosis or occlusion: a systematic review and Meta-analysis. Stroke 2012; 43: 2884–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito M, Lammertsma AA, Wise RJS, et al. Measurement of regional cerebral blood flow and oxygen utilisation in patients with cerebral tumours using 15O and positron emission tomography: Analytical techniques and preliminary results. Neuroradiology 1982; 23: 63–74. [DOI] [PubMed] [Google Scholar]
- 6.Ishii K, Kitagaki H, Kono M, et al. Decreased medial temporal oxygen metabolism in Alzheimer’s disease shown by PET. J Nucl Med 1996; 37: 1159–1165. [PubMed] [Google Scholar]
- 7.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. Stroke 2005; 36: e83–e99. [DOI] [PubMed] [Google Scholar]
- 8.Mintun MA, Raichle ME, Martin WR, et al. Brain oxygen utilization measured with O-15 radiotracers and positron emission tomography. J Nucl Med 1984; 25: 177–187. [PubMed] [Google Scholar]
- 9.Herscovitch P, Markham J, Raichle ME.Brain blood flow measured with intravenous H2(15)O. I. Theory and error analysis. J Nucl Med 1983; 24: 782–789. [PubMed] [Google Scholar]
- 10.Ibaraki M, Miura S, Shimosegawa E, et al. Quantification of cerebral blood flow and oxygen metabolism with 3-dimensional PET and 15O: validation by comparison with 2-dimensional PET. J Nucl Med 2008; 49: 50–59. [DOI] [PubMed] [Google Scholar]
- 11.Ibaraki M, Shimosegawa E, Miura S, et al. PET measurements of CBF, OEF, and CMRO2 without arterial sampling in hyperacute ischemic stroke: method and error analysis. Ann Nucl Med 2004; 18: 35–44. [DOI] [PubMed] [Google Scholar]
- 12.Yamauchi H, Fukuyama H, Nagahama Y, et al. Evidence of misery perfusion and risk for recurrent stroke in major cerebral arterial occlusive diseases from PET. J Neurol Neurosurg Psychiatry 1996; 61: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derdeyn CP, Videen TO, Simmons NR, et al. Count-based PET method for predicting ischemic stroke in patients with symptomatic carotid arterial occlusion. Radiology 1999; 212: 499–506. [DOI] [PubMed] [Google Scholar]
- 14.Powers WJ, Zazulia AR.PET in cerebrovascular disease. PET Clin 2010; 5: 83106–83106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo K, Liu T, Murakami T, et al. Oxygen extraction fraction measurement using quantitative susceptibility mapping: comparison with positron emission tomography. J Cereb Blood Flow Metab 2016; 36: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powers WJ, Videen TO, Markham J, et al. Selective defect of in vivo gliycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A 2007; 104: 2945–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y, Vlassenko AG, Couture LE, et al. Quantitative hemodynamic PET imaging using image-derived arterial input function and a PET/MR hybrid scanner. J Cereb Blood Flow Metab 2017; 37: 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y, Arbelaez AM, Benzinger TL, et al. Noninvasive estimation of the arterial input function in positron emission tomography imaging of cerebral blood flow. J Cereb Blood Flow Metab 2013; 33: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z, Sur S, Soldan A, et al. Brain oxygen extraction by using MRI in older individuals: relationship to apolipoprotein E genotype and amyloid burden. Radiology 2019; 292: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu H, Xu F, Grgac K, et al. Calibration and validation of TRUST MRI for the estimation of cerebral blood oxygenation. Magn Reson Med 2012; 67: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H, Ge Y.Quantitative evaluation of oxygenation in venous vessels using T2-relaxation-under-spin-tagging MRI. Magn Reson Med 2008; 60: 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X, Yablonskiy DA.Quantitative BOLD: mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: default state. Magn Reson Med 2007; 57: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yablonskiy DA, Sukstanskii AL, He X.BOLD-based techniques for quantifying brain hemodynamic and metabolic properties – theoretical models and experimental approaches. NMR Biomed 2013; 26: 963–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An H, Liu Q, Chen Y, et al. Evaluation of MR-Derived cerebral oxygen metabolic index in experimental hyperoxic hypercapnia, hypoxia, and ischemia. Stroke 2009; 40: 2165–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An H, Lin W.Impact of intravascular signal on quantitative measures of cerebral oxygen extraction and blood volume under normo- and hypercapnic conditions using an asymmetric spin echo approach. Magn Reson Med 2003; 50: 708–716. [DOI] [PubMed] [Google Scholar]
- 26.An H, Lin W.Quantitative measurements of cerebral blood oxygen saturation using magnetic resonance imaging. J Cereb Blood Flow Metab 2000; 20: 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolar DS, Rosen BR, Sorensen A, et al. QUantitative imaging of eXtraction of oxygen and TIssue consumption (QUIXOTIC) using venular-targeted velocity-selective spin labeling. Magn Reson Med 2011; 66: 1550–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis TL, Kwong KK, Weisskoff RM, et al. Calibrated functional MRI: mapping the dynamics of oxidative metabolism. Proc Natl Acad Sci U S A 1998; 95: 1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoge RD.Calibrated FMRI. NeuroImage 2012; 62: 930–937. [DOI] [PubMed] [Google Scholar]
- 30.Gauthier CJ, Hoge RD.Magnetic resonance imaging of resting OEF and CMRO2 using a generalized calibration model for hypercapnia and hyperoxia. NeuroImage 2012; 60: 1212–1225. [DOI] [PubMed] [Google Scholar]
- 31.Blockley NP, Griffeth VE, Simon AB, et al. A review of calibrated blood oxygenation level-dependent (BOLD) methods for the measurement of task-induced changes in brain oxygen metabolism. NMR Biomed 2013; 26: 987–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain V, Langham MC, Wehrli FW.MRI estimation of global brain oxygen consumption rate. J Cereb Blood Flow Metab 2010; 30: 1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wehrli FW, Fan AP, Rodgers ZB, et al. Susceptibility-based time-resolved whole-organ and regional tissue oximetry. NMR Biomed 2017; 30: e3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Rochefort L, Liu T, Kressler B, et al. Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging. Magn Reson Med 2010; 63: 194–206. [DOI] [PubMed] [Google Scholar]
- 35.Fan AP, Benner T, Bolar DS, et al. Phase-based regional oxygen metabolism (PROM) using MRI. Magn Reson Med 2012; 67: 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan AP, Bilgic B, Gagnon L, et al. Quantitative oxygenation venography from MRI phase. Magn Reson Med 2014; 72: 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Liu T, Gupta A, et al. Quantitative mapping of cerebral metabolic rate of oxygen (CMRO2) using quantitative susceptibility mapping (QSM). Magn Reson Med 2015; 74: 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Cho J, Zhou D, et al. Quantitative susceptibility mapping-based cerebral metabolic rate of oxygen mapping with minimum local variance. Magn Reson Med 2018; 79: 172–179. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Zhou D, Nguyen TD, et al. Cerebral metabolic rate of oxygen (CMRO2) mapping with hyperventilation challenge using quantitative susceptibility mapping (QSM). Magn Reson Med 2017; 77: 1762–1773. [DOI] [PubMed] [Google Scholar]
- 40.Cho J, Kee Y, Spincemaille P, et al. Cerebral metabolic rate of oxygen (CMRO2) mapping by combining quantitative susceptibility mapping (QSM) and quantitative blood oxygenation level-dependent imaging (qBOLD). Magn Reson Med 2018; 80: 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho J, Zhang S, Kee Y, et al. Cluster analysis of time evolution (CAT) for quantitative susceptibility mapping (QSM) and quantitative blood oxygen level-dependent magnitude (qBOLD)-based oxygen extraction fraction (OEF) and cerebral metabolic rate of oxygen (CMRO2) mapping. Magn Reson Med 2020; 83: 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgos N, Cardoso MJ, Thielemans K, et al. Attenuation correction synthesis for hybrid PET-MR scanners: application to brain studies. IEEE Trans Med Imaging 2014; 33: 2332–2341. [DOI] [PubMed] [Google Scholar]
- 43.Markiewicz PJ, Thielemans K, Schott JM, et al. Rapid processing of PET list-mode data for efficient uncertainty estimation and data analysis. Phys Med Biol 2016; 61: 322–336. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Cho J, Zhou D, et al. Quantitative susceptibility mapping-based cerebral metabolic rate of oxygen mapping with minimum local variance. Magn Reson Med 2017; 79: 172–179. [DOI] [PubMed]
- 45.Yablonskiy DA, Haacke EM.Theory of NMR signal behavior in magnetically inhomogeneous tissues: the static dephasing regime. Magn Reson Med 1994; 32: 749–763. [DOI] [PubMed] [Google Scholar]
- 46.Liu T, Wisnieff C, Lou M, et al. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med 2013; 69: 467–476. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, Khalidov I, de Rochefort L, et al. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed 2011; 24: 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Liu T, de Rochefort L, et al. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. NeuroImage 2012; 59: 2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Liu T.Quantitative susceptibility mapping (QSM): decoding MRI data for a tissue magnetic biomarker. Magn Reson Med 2015; 73: 82–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Kee Y, Zhou D, et al. Preconditioned total field inversion (TFI) method for quantitative susceptibility mapping. Magn Reson Med 2017; 78: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Spincemaille P, Yao Y, et al. MEDI+0: morphology enabled dipole inversion with automatic uniform cerebrospinal fluid zero reference for quantitative susceptibility mapping. Magn Reson Med 2018; 79: 2795–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenkinson M, Smith S.A global optimisation method for robust affine registration of brain images. Med Image Anal 2001; 5: 143–156. [DOI] [PubMed] [Google Scholar]
- 53.Jenkinson M, Bannister P, Brady M, et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 2002; 17: 825–841. [DOI] [PubMed] [Google Scholar]
- 54.Hajnal JV, Saeed N, Soar EJ, et al. A registration and interpolation procedure for subvoxel matching of serially acquired MR images. J Comput Assist Tomogr 1995; 19: 289–296. [DOI] [PubMed] [Google Scholar]
- 55.Rowland DJ, Garbow JR, Laforest R, et al. Registration of [18F]FDG microPET and small-animal MRI. Nucl Med Biol 2005; 32: 567–572. [DOI] [PubMed] [Google Scholar]
- 56.Joshi A, Koeppe RA, Fessler JA.Reducing between scanner differences in multi-center PET studies. NeuroImage 2009; 46: 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33: 341–355. [DOI] [PubMed] [Google Scholar]
- 58.Su Y, Blazey TM, Snyder AZ, et al. Quantitative amyloid imaging using image-derived arterial input function. PLoS One 2015; 10: e0122920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson DL, Noble JA.An adaptive segmentation algorithm for time-of-flight MRA data. IEEE Trans Med Imaging 1999; 18: 938–945. [DOI] [PubMed] [Google Scholar]
- 60.Raichle ME, Martin WR, Herscovitch P, et al. Brain blood flow measured with intravenous H2(15)O. II. Implementation and validation. J Nucl Med 1983; 24: 790–798. [PubMed] [Google Scholar]
- 61.Grubb RL, Jr, Raichle ME, Higgins CS, et al. Measurement of regional cerebral blood volume by emission tomography. Ann Neurol 1978; 4: 322–328. [DOI] [PubMed] [Google Scholar]
- 62.Bland JM, Altman DG.Measuring agreement in method comparison studies. Stat Methods Med Res 1999; 8: 135–160. [DOI] [PubMed] [Google Scholar]
- 63.Baron JC.Mapping the ischaemic penumbra with PET: implications for acute stroke treatment. Cerebrovasc Dis 1999; 9: 193–201. [DOI] [PubMed] [Google Scholar]
- 64.Sun X, He G, Qing H, et al. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A 2006; 103: 18727–18732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carvalho C, Correia SC, Santos RX, et al. Role of mitochondrial-mediated signaling pathways in Alzheimer disease and hypoxia. J Bioenerg Biomembr 2009; 41: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapp BD, Stys PK.Virtual hypoxia and chronic necrosis of demyelinated axons in multiple sclerosis. Lancet Neurol 2009; 8: 280–291. [DOI] [PubMed] [Google Scholar]
- 67.Jones TH, Morawetz RB, Crowell RM, et al. Thresholds of focal cerebral ischemia in awake monkeys. J Neurosurg 1981; 54: 773–782. [DOI] [PubMed] [Google Scholar]
- 68.Davis SM, Donnan GA, Parsons MW, et al. Effects of alteplase beyond 3 h after stroke in the echoplanar imaging thrombolytic evaluation trial (EPITHET): a placebo-controlled randomised trial. Lancet Neurol 2008; 7: 299–309. [DOI] [PubMed] [Google Scholar]
- 69.Hacke W, Furlan AJ, Al-Rawi Y, et al. Intravenous desmoteplase in patients with acute ischaemic stroke selected by MRI perfusion-diffusion weighted imaging or perfusion CT (DIAS-2): a prospective, randomised, double-blind, placebo-controlled study. Lancet Neurol 2009; 8: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furlan AJ, Eyding D, Albers GW, DEDAS Investigators et al. Dose escalation of desmoteplase for acute ischemic stroke (DEDAS): evidence of safety and efficacy 3 to 9 hours after stroke onset. Stroke 2006; 37: 1227–1231. [DOI] [PubMed] [Google Scholar]
- 71.Vlassenko AG, Vaishnavi SN, Couture L, et al. Spatial correlation between brain aerobic glycolysis and amyloid-beta (abeta) deposition. Proc Natl Acad Sci U S A 2010; 107: 17763–17767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raichle ME, MacLeod AM, Snyder AZ, et al. A default mode of brain function. Proc Natl Acad Sci U S A 2001; 98: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carpenter DA, Grubb RL, Jr, Tempel LW, et al. Cerebral oxygen metabolism after aneurysmal subarachnoid hemorrhage. J Cereb Blood Flow Metab 1991; 11: 837–844. [DOI] [PubMed] [Google Scholar]
- 74.Ma Y, Sun H, Cho J, et al. Cerebral OEF quantification: a comparison study between quantitative susceptibility mapping and dual-gas calibrated BOLD imaging. Magn Reson Med 2020; 83: 68–82. [DOI] [PubMed] [Google Scholar]
- 75.Bulte DP, Kelly M, Germuska M, et al. Quantitative measurement of cerebral physiology using respiratory-calibrated MRI. NeuroImage 2012; 60: 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wise RG, Harris AD, Stone AJ, et al. Measurement of OEF and absolute CMRO2: MRI-based methods using interleaved and combined hypercapnia and hyperoxia. NeuroImage 2013; 83: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Germuska M, Bulte DP.MRI measurement of oxygen extraction fraction, mean vessel size and cerebral blood volume using serial hyperoxia and hypercapnia. NeuroImage 2014; 92: 132–142. [DOI] [PubMed] [Google Scholar]
- 78.Fan AP, Schäfer A, Huber L, et al. Baseline oxygenation in the brain: correlation between respiratory-calibration and susceptibility methods. NeuroImage 2016; 125: 920–931. [DOI] [PubMed] [Google Scholar]
- 79.Germuska M, Merola A, Murphy K, et al. A forward modelling approach for the estimation of oxygen extraction fraction by calibrated fMRI. NeuroImage 2016; 139: 313–323. [DOI] [PubMed] [Google Scholar]
- 80.Lajoie I, Tancredi FB, Hoge RD.Regional reproducibility of BOLD calibration parameter M, OEF and resting-state CMRO2 measurements with QUO2 MRI. PLoS One 2016; 11: e0163071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Delso G, Furst S, Jakoby B, et al. Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. J Nucl Med 2011; 52: 1914–1922. [DOI] [PubMed] [Google Scholar]
- 82.Su Y, Rubin BB, McConathy J, et al. Impact of MR-based attenuation correction on neurologic PET studies. J Nucl Med 2016; 57: 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan AP, Khalil AA, Fiebach JB, et al. Elevated brain oxygen extraction fraction measured by MRI susceptibility relates to perfusion status in acute ischemic stroke. J Cereb Blood Flow Metab 2020; 40: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X20973951 for Cerebral oxygen extraction fraction (OEF): Comparison of challenge-free gradient echo QSM+qBOLD (QQ) with 15O PET in healthy adults by Junghun Cho, John Lee, Hongyu An, Manu S Goyal, Yi Su and Yi Wang in Journal of Cerebral Blood Flow & Metabolism