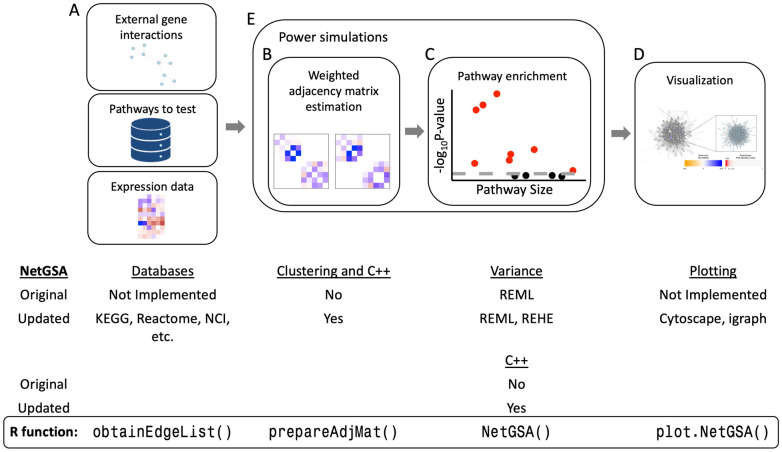
Fig 1. NetGSA methodology highlighting package updates.
(A) NetGSA takes external gene interactions, a matrix of pathways to test, and expression data as inputs. The new netgsa incorporates existing external databases such as KEGG, Reactome, NCI, etc. (B) Weighted adjacency matrices are calculated for each condition (two conditions shown). Users now have the option to use clustering in calculating the weighted adjacency matrices. (C) Pathway enrichment is performed using weighted adjacency matrices. In addition to REML, users can estimate the variance parameters using REHE. Faster matrix calculations are also incorporated in C++ (B & C). (D) Visualization is now available with the option to use either Cytoscape or igraph. (E) Power simulations involved steps (B) and (C).